Protein ubiquitination plays an essential role in a variety of cellular signaling pathways [1, 2]. Ubiquitin (Ub) can form eight types of ubiquitin chains through its own seven lysine residues and an N-terminal amino group. Different ubiquitin chains adopt different conformations, and therefore encode distinct information [3, 4]. For instance, K48-linked and K63-linked ubiquitin chains acting as degradation and non-degradation signal, respectively [5, 6], while other atypical ubiquitin chains play important roles in the processes of anti-virus [7-10], protein trafficking [11]. Recent studies have shown that in addition to homogeneously linked ubiquitin chains, mixed or branched ubiquitin chains also control various cellular activities [12-16]. For example, K29, 48-branched ubiquitin chains enhance substrate degradation by proteasome [12]. In the NF-kB signaling pathway, the K48, 63-branched ubiquitin chains can be recognized normally by the reader protein TAB 2, but stay inert towards deubiquitinase (DUB) CYLD. Therefore, its stability against hydrolysis significantly enhances its activation effect of the NF-kB signal [13]. Recently, Rape et al. reported that K11, 48-branched ubiquitin chains can act as an enhanced signal during eukaryotic mitosis and protein quality control to ensure the rapid degradation of cell-cycle proteins and misfolded proteins by proteasome [14, 15]. These results suggest that branched ubiquitin chains tune the readout of the ubiquitin code in a sophisticated manner. However, due to the lack of linkage- and length-defined branched ubiquitin chains in vitro, those linkage- and length-defined K11, 48-branched ubiquitin chains remain elusive with regard to their biochemical properties such as their binding affinity and enzymolysis stability.
Herein, we employed chemically synthesized K11, 48-branched ubiquitin chains to determine their binding affinity with the proteasome ubiquitin receptor S5a by using Surface Plasma Resonance (SPR) technique. Our results revealed that S5a bounds to K48-linked, K11, 48-branched and K11-linked chains in decreasing order. Moreover, we carried out deubiquitination experiments with proteasome deubiquitinase Rpn11-containing complex (Rpn8/11, lid-core, lid). We found that the Rpn11 preferably hydrolyzed branched ubiquitin chains than K11 or K48 linked ubiquitin chains, which exhibited a unique hydrolysis properties of K11, 48-branched ubiquitin chains.
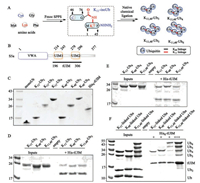
Chemical protein synthesis [17-40] allows the synthesis of ubiquitin chains with defined chemical structure, and greatly contributed to our understanding of the biochemical and structural properties of ubiquitin chains. To study the binding properties of K11, 48-branched ubiquitin chains towards ubiquitin binding proteins, K11, 48-branched tri- to hexa-ubiquitin were chemically synthesized as previously reported [41]. Briefly, a K11-isoUb unit was premade via 9-fluorenylmethoxycarbonyl solid-phase peptide synthesis (Fmoc-SPPS), then K11, 48-branched ubiquitin chains were assembled from K11-isoUb via classical native chemical ligation (NCL) (Fig. 1A). S5a is an essential proteasome ubiquitin receptor, using its C-terminal tandem ubiquitin interacting motifs (tUIM) to bind polyubiquitin chains (Fig. 1B) [42-44]. We therefore recombinant expressed N-terminal His6 tagged tUIM for pulldown assay and SPR assay. We also prepared the K11 and K48-linked di- to tetra-Ub by enzymatic approach as previously reported (Fig. 1C) [45].
|
Download:
|
| Fig. 1. His6-based pulldown assays. (A) Branched Ub chains synthesized via IsoUb strategy. (B) Domain structure of S5a. (C) SDS-PAGE of His-tUIM and enzymatic synthesized Ub chains. (D) Tri-Ub pulldown. (E) Tetra-Ub pulldown. (F) Competitive pulldown assays. | |
With ubiquitin chains and tUIM in hand, we carried out His6-tag based pulldown experiment to exam the binding properties of different ubiquitin towards tUIM. We first performed pulldown assay using K11- and K48-linked di-Ub (Ub2). K11- or K48-linked Ub2 were incubated with tUIM (immobilized on Ni-NTA resin) for 2 h. After elution with imidazole, the eluate was determined by SDS-PAGE. Both K11- and K48-linked Ub2 cannot be detected by Coomassie brilliant blue (CBB) staining (data not shown), indicating that Ub2 is not an effective degradation signal. We then performed pulldown assay using tri-Ub (Ub3), and we found that K11-, K48- and K11, 48-linked Ub3 can be recognized by tUIM (Fig. 1D). K11-linked Ub3 showed a weaker binding than K48- or K11, 48-linked Ub3. Similarly, pulldown assay with tetra-Ub (Ub4) showed that tUIM could recognize all three types of ubiquitin chains (Fig. 1E). When compared to Ub3, Ub4 has a stronger interaction with tUIM, suggesting that longer ubiquitin chain is an effective degradation signal.
To further certify above results, we carried out competitive pulldown experiments. When equal mass of Ub, K11-linked Ub2, Ub3 and Ub4 were incubated with tUIM separately, we found that tUIM preferentially bound to Ub4 than other shorter ubiquitin chains, although the short ubiquitin chains have higher concentrations (Fig. 1F, lane 6). Similarly, we carried out pulldown experiment using K48-linked ubiquitin chains, and found that tUIM mainly bound to K48-linked Ub4, as well as a small amount of Ub3 (Fig. 1F, lane 7). Di-Ub and mono-Ub cannot be detected by CBB staining. When we applied to K11, 48-branched ubiquitin chains, we found that tUIM mainly bound to hexa-Ub (Ub6) and a small number of penta-Ub (Ub5) and tetra-Ub (Ub4) (Fig. 1F, lane 8). When we increased the amount of tUIM to the sum of all the amount of ubiquitin chains, Ub4 and Ub3 could be pulled out, while the Ub2 (K11-linked) was still undetectable by CBB staining (Fig. 1F, lane 9). Above experiments showed that in consist with previous reports, tUIM preferred associating with longer ubiquitin chains [46], which sequentially meant that longer ubiquitin chains were more efficient proteasome targeting signals than shorter chains [47].
SPR technique has been widely used to determine the affinity of ubiquitin receptor to polyubiquitin chains [48]. To quantitatively compare the interaction between S5a and different conjugated chains, we measured the affinity of different Ub chains to tUIM by SPR. The SPR results were summarized in Table 1. K11- and K48-linked Ubn (n = 2, 3, 4) and K11, 48-branched Ubn (n = 3, 4, 5, 6) showed a significant increase in affinity with increasing chain length for all types of chains, explaining the preferred binding to longer chains, agreeing with the observation in the above pulldown assays. However, tUIM bound to K48-linked chains with the highest affinity when compared with K11-linked or K11, 48-branched chains of equal molecular mass, which is also in consist with previous reports [49]. As shown in Fig. 2C, the affinity of K11, 48-Ub3 was lower than K48-Ub3 (Fig. 2B), but higher than K11-Ub3 (Fig. 2A). Moreover, the measured dissociation constant (Kd) of K11, 48-Ub3 (7.1 mmol/L) was approximately in the middle between that of K48-Ub3 (4.0 mmol/L, 1.8-fold decrease) and K11-Ub3 (17.1 mmol/L, 2.4-fold increase), while Kd of K11, 48-Ub4 (Fig. 2F, 4.2 mmol/L) was closer to that of K11-Ub4 (Fig. 2D, 5.0 mmol/L, 1.2-fold increase) than K48-Ub4 (Fig. 2E, 1.2 mmol/L, 3.5-fold decrease). Thus, these experiments showed that tUIM preferred binding to K48-linked ubiquitin chains than synthetic K11, 48-branched or K11-linked chains, and the affinity of branched chains was closer to the K11 chains with the increase of the chain length.
|
|
Table 1 Dissociation constant (KD, mmol/L) of ubiquitin chains to tUIM. |

|
Download:
|
| Fig. 2. Quantitative determination the affinity of tUIM to ubiquitin chains. SPR responses and fitting curves for K11-Ub3 (A), K48-Ub3 (B), K11,48-Ub3 (C), K11-Ub4 (D), K48-Ub4 (E), and K11,48-Ub4 (F). | |
To assess the disassembly properties of K11, 48-branched ubiquitin chains, we performed deubiquitination assays using yeast proteasome-associated deubiquitinase Rpn11-containing complex Rpn8/11, lid-core (Rpn5/6/8/9/11), lid (Rpn3/5/6/7/8/9/ 11/12/15) (Fig. S2A in Supporting information). Rpn8/11 works together with other DUBs to maintain the pool of free Ub (in living cells) [50, 51]. We incubated the ubiquitin chains with purified Rpn11-containing complex (Rpn8/11, lid-core, lid) and monitored the cleavage reaction by CBB or blotting with anti-Ub. Purified truncated Rpn11(1-239) alone showed no activity towards K11- or K48-linked Ub2 (Fig. S2C in Supporting information), which is consistent with previous studies that Rpn11's DUB activity is dependent on the interaction with Rpn8 [52, 53]. When K11- or K48-linked Ub2 were incubated with C-terminal truncated Rpn8 (1- 178)-Rpn11 (1-239) heterodimer, after 60 min, about 60% of K11- and 30% of K48-linked Ub2 were cleaved (Fig. S2D in Supporting information), which is similar to the reported results [52]. However, cleavage of K11 or K48-linked Ub3 and Ub4 showed much slower than the corresponding Ub2, as shown in Figs. 3A and C, less than 10% of K11- and K48-linked Ub3 or Ub4 can be cleaved by Rpn8 (1-178)-Rpn11 (1-239) heterodimer after 6 h, despite that longer ubiquitin chains potentially had more binding sites on a per mole basis. Those longer K11 and K48-linked polyubiquitins may adopt more closed conformation, as found in K48-Ub4 [54]. The closed conformations slow down the cleavage of isopeptide bond. Surprisingly, Rpn8-Rpn11 heterodimer showed significantly higher activity for branched structures (Figs. 3A and C). Almost all of the K11, 48-Ub3 and K11, 48-Ub4 were hydrolyzed by Rpn8-Rpn11 heterodimer after 6 h (Figs. 3A and C). MS/MS analysis demonstrated that Rpn8-Rpn11 heterodimer cleaved both K11 and K48 linkages (Fig. S2B in Supporting information). Similar preference for branched chains was also observed when the five-component proteasomal lid-core (Figs. S2E and F in Supporting information) and the nine-component lid sub-complex (Figs. 3B and D, and Fig. S2G in Supporting information) were applied except that the efficiency was lower than Rpn8/11, probably due to the steric inhibition of active site by Rpn5 [55]. Moreover, Rpn8/11 gradually trimmed K11, 48-Ub5 and K11, 48-Ub6 with similar efficiency of K11, 48-Ub3 or K11, 48-Ub4 (left lane in Figs. 3E and F), revealing that Rpn8/11 could efficiently target the internal K11-linked Ub3 in branched Ub5 or Ub6. These results indicated that branched ubiquitin chains might adopt more open conformations to facilitate the isopeptide bond cleavage by Rpn11-containg complex. Therefore, using the length and linkage-defined branched ubiquitin chain system, we revealed that synthetic K11, 48-branched ubiquitin chains were preferentially hydrolyzed by proteasome DUB Rpn11. Taken together, our data suggest that the effective binding of K11, 48-branched chains to proteasomal ubiquitin receptors and the hydrolysis preference for proteasomal DUB might collectively contribute to the enhanced degradation of cell-cycle regulators (Fig. 4).

|
Download:
|
| Fig. 3. Disassembly of Ub chains with Rpn11-containing complex. Disassembly of differently linked triUb by (A) Rpn8-Rpn11 heterodimer and (B) proteasomal lid; Hydrolysis of tetraUb with different linkages by (C) Rpn8-Rpn11 heterodimer and (D) proteasomal lid; Rpn11-containing complex trimmed branched (E) penta- and (F) hexa-ubiquitin chains. Experiments with Rpn8-Rpn11 heterodimer were analyzed by SDS-PAGE, and results of lid-core and lid were analyzed by western blotting using anti-Ub (Abcam). The asterisk refers Rpn8-Rpn11 dimer. | |

|
Download:
|
| Fig. 4. The proposed mechanism for enhanced substrate degradation by branched ubiquitin chains. | |
In conclusion, we characterized the binding and hydrolysis properties of the K11, 48-branched ubiquitin chains for the first time. The binding experiments with S5a showed that the affinity of the branched ubiquitin chains was stronger than K11-linked chains. Deubiquitination assays revealed that the K11, 48-branched chains could be more efficiently hydrolyzed by Rpn11 when compared with the homotypic K11 or K48 ubiquitin chain, suggesting that the branched ubiquitin chains might adopt an unique conformation that could be more effectively recognized by Rpn11.
AcknowledgmentsWe thank Prof. Ziqing Mei (Chinese Academy of Agricultural Sciences, Beijing) for generously providing plasmids of yeast 26S proteasome-associated deubiquitinase Rpn11-containing complex. This work was supported by the National Natural Science Foundation of China (Nos. U1732161, 91753120).
Appendix A. Supplementary dataSupplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.cclet.2018.03.022.
| [1] |
D. Komander, M. Rape, Annu. Rev. Biochem. 81 (2012) 203-229. DOI:10.1146/annurev-biochem-060310-170328 |
| [2] |
R. Yau, M. Rape, Nat. Cell Biol. 18 (2016) 579-586. DOI:10.1038/ncb3358 |
| [3] |
Y. Kulathu, D. Komander, Nat. Rev. Mol. Cell Biol. 13 (2012) 508-523. DOI:10.1038/nrm3394 |
| [4] |
K.N. Swatek, D. Komander, Cell Res. 26 (2016) 399-422. DOI:10.1038/cr.2016.39 |
| [5] |
M. Hochstrasser, Annu. Rev. Genet. 30 (1996) 405-439. DOI:10.1146/annurev.genet.30.1.405 |
| [6] |
Z.J. Chen, L.J. Sun, Mol. Cell 33 (2009) 275-286. DOI:10.1016/j.molcel.2009.01.014 |
| [7] |
Q. Wang, X. Liu, Y. Cui, et al., Immunity 41 (2014) 919-933. DOI:10.1016/j.immuni.2014.11.011 |
| [8] |
H. Liu, L. Zhang, J. Sun, et al., J. Virol. 91 (2017) e02234-16. |
| [9] |
K.M. Sparrer, S. Gableske, M.A. Zurenski, et al., Nat.Microbiol. 2 (2017) 1543-1557. DOI:10.1038/s41564-017-0017-2 |
| [10] |
J. Liu, C. Han, B. Xie, et al., Nat. Immunol. 15 (2014) 612-622. DOI:10.1038/ni.2898 |
| [11] |
W.C. Yuan, Y.R. Lee, S.Y. Lin, et al., Mol. Cell 54 (2014) 586-600. DOI:10.1016/j.molcel.2014.03.035 |
| [12] |
C. Liu, W. Liu, Y. Ye, W Li, Nat. Commun 8 (2017) 14274. DOI:10.1038/ncomms14274 |
| [13] |
F. Ohtake, Y. Saeki, S. Ishido, J. Kanno, K. Tanaka, Mol. Cell 64 (2016) 251-266. DOI:10.1016/j.molcel.2016.09.014 |
| [14] |
H.J. Meyer, M. Rape, Cell 157 (2014) 910-921. DOI:10.1016/j.cell.2014.03.037 |
| [15] |
R.G. Yau, K. Doerner, E.R. Castellanos, et al., Cell 171 (2017) 918-933. DOI:10.1016/j.cell.2017.09.040 |
| [16] |
C.H. Emmerich, A. Ordureau, S. Strickson, et al., Proc. Natl. Acad. Sci. U. S. A. 110 (2013) 15247-15252. DOI:10.1073/pnas.1314715110 |
| [17] |
H.X. Li, S.W. Dong, Sci. China Chem. 60 (2017) 201-213. |
| [18] |
Y. Gui, L.Q. Qiu, Y.H. Li, et al., J. Am. Chem. Soc. 138 (2016) 4890-4899. DOI:10.1021/jacs.6b01202 |
| [19] |
C.L. Lee, X.C. Li, Sci. China Chem. 59 (2016) 1061-1064. |
| [20] |
K. Jin, T.L. Li, H.Y. Chow, et al., Angew. Chem. Int. Ed. 56 (2017) 14607-14611. DOI:10.1002/anie.201709097 |
| [21] |
C.L. Lee, H. Liu, C.T.T. Wong, et al., J. Am. Chem. Soc. 138 (2016) 10477-10484. DOI:10.1021/jacs.6b04238 |
| [22] |
K. Jin, L.H. Sam, K.H.L. Po, et al., Nat. Commun 7 (2016) 12394. DOI:10.1038/ncomms12394 |
| [23] |
Q.Q. He, J.B. Li, Y.K. Qi, et al., Sci. China Chem. 60 (2017) 621-627. DOI:10.1007/s11426-016-0386-4 |
| [24] |
K. Ajish Kumar, M. Haj-Yahya, D. Olschewski, et al., Angew. Chem. Int. Ed. 48 (2009) 8090-8094. DOI:10.1002/anie.v48:43 |
| [25] |
R. Yang, K.K. Pasunooti, F. Li, et al., J. Am. Chem. Soc. 131 (2009) 13592-13593. DOI:10.1021/ja905491p |
| [26] |
F. El Oualid, R. Merkx, R. Ekkebus, et al., Angew. Chem. Int. Ed. 49 (2010) 10149-10153. DOI:10.1002/anie.201005995 |
| [27] |
K. Kumar, S.N. Bavikar, L. Spasser, et al., Angew. Chem. Int. Ed. 50 (2011) 6137-6141. DOI:10.1002/anie.v50.27 |
| [28] |
G.M. Fang, Y.M. Li, F. Shen, et al., Angew. Chem. Int. Ed. 50 (2011) 7645-7649. DOI:10.1002/anie.201100996 |
| [29] |
G.M. Fang, J.X. Wang, L. Liu, Angew. Chem. Int. Ed. 51 (2012) 10347-10350. DOI:10.1002/anie.201203843 |
| [30] |
M. Haj-Yahya, B. Fauvet, Y. Herman-Bachinsky, et al., Proc. Natl. Acad. Sci. U. S. A. 110 (2013) 17726-17731. DOI:10.1073/pnas.1315654110 |
| [31] |
J.X. Wang, G.M. Fang, Y. He, et al., Angew. Chem. Int. Ed. 54 (2015) 2194-2198. DOI:10.1002/anie.201408078 |
| [32] |
Y.C. Huang, Y.M. Li, Y. Chen, et al., Angew. Chem. Int. Ed. 52 (2013) 4858-4862. DOI:10.1002/anie.v52.18 |
| [33] |
Y.M. Li, Y.T. Li, M. Pan, et al., Angew. Chem. Int. Ed. 53 (2014) 2198-2202. DOI:10.1002/anie.201310010 |
| [34] |
S. Tang, Y.Y. Si, Z.P. Wang, et al., Angew. Chem. Int. Ed. 54 (2015) 5713-5717. DOI:10.1002/anie.201500051 |
| [35] |
M. Pan, S. Gao, Y. Zheng, et al., J. Am. Chem. Soc. 138 (2016) 7429-7435. DOI:10.1021/jacs.6b04031 |
| [36] |
S. Gao, M. Pan, Y. Zheng, et al., J. Am. Chem. Soc. 138 (2016) 14497-14502. DOI:10.1021/jacs.6b09545 |
| [37] |
J. Liang, L. Zhang, X.L. Tan, et al., Angew. Chem. Int. Ed. 56 (2017) 2744-2748. DOI:10.1002/anie.201611659 |
| [38] |
J. Li, Q. He, Y. Liu, et al., ChemBioChem 18 (2017) 176-180. DOI:10.1002/cbic.201600551 |
| [39] |
J.S. Zheng, S. Tang, Y.K. Qi, et al., Nat. Protoc. 8 (2013) 2483-2495. DOI:10.1038/nprot.2013.152 |
| [40] |
Z. Wang, W. Xu, L. Liu, et al., Nat. Chem. 8 (2016) 698-704. DOI:10.1038/nchem.2517 |
| [41] |
S. Tang, L.J. Liang, Y.Y. Si, et al., Angew. Chem. Int. Ed. 56 (2017) 13333-13337. DOI:10.1002/anie.201708067 |
| [42] |
Q. Deveraux, V. Ustrell, C. Pickart, et al., J. Biol. Chem. 269 (1994) 7059-7061. |
| [43] |
P. Young, Q. Deveraux, R.E. Beal, et al., J. Biol. Chem. 273 (1998) 5461-5467. DOI:10.1074/jbc.273.10.5461 |
| [44] |
N. Zhang, Q. Wang, A. Ehlinger, et al., Mol. Cell 35 (2009) 280-290. DOI:10.1016/j.molcel.2009.06.010 |
| [45] |
K.C. Dong, E. Helgason, C. Yu, et al., Structure 19 (2011) 1053-1063. DOI:10.1016/j.str.2011.06.010 |
| [46] |
Q. Wang, P. Young, K.J. Walters, J. Mol. Biol. 348 (2005) 727-739. DOI:10.1016/j.jmb.2005.03.007 |
| [47] |
J.S. Thrower, L. Hoffman, M. Rechsteiner, et al., EMBO J. 19 (2000) 94-102. DOI:10.1093/emboj/19.1.94 |
| [48] |
S. Raasi, I. Orlov, K.G. Fleming, et al., J. Mol. Biol. 341 (2004) 1367-1379. DOI:10.1016/j.jmb.2004.06.057 |
| [49] |
G.L. Grice, I.T. Lobb, M.P. Weekes, et al., Cell Rep. 12 (2015) 545-553. DOI:10.1016/j.celrep.2015.06.061 |
| [50] |
R. Verma, L. Aravind, R. Oania, et al., Science 298 (2002) 611-615. DOI:10.1126/science.1075898 |
| [51] |
T. Yao, R.E. Cohen, Nature 419 (2002) 403-407. DOI:10.1038/nature01071 |
| [52] |
E.J. Worden, C. Padovani, A. Martin, Nat. Struct. Mol. Biol. 21 (2014) 220-227. DOI:10.1038/nsmb.2771 |
| [53] |
G.R. Pathare, I. Nagy, P. Sled z, et al., Proc. Natl. Acad. Sci. U. S. A. 111 (2014) 2984-2989. DOI:10.1073/pnas.1400546111 |
| [54] |
M.J. Eddins, R. Varadan, D. Fushman, et al., J. Mol. Biol. 367 (2007) 204-211. DOI:10.1016/j.jmb.2006.12.065 |
| [55] |
C.M. Dambacher, E.J. Worden, M.A. Herzik Jr, et al., eLife 5 (2016) e13027. |
 2018, Vol. 29
2018, Vol. 29 



