b Key Laboratory of Biochip Technology, Biotech and Health Centre, Shenzhen Research Institute of City University of Hong Kong, Shenzhen 518057, China;
c Department of Chemistry, The Chinese University of Hong Kong, Hong Kong, China;
d College of Biotechnology and bioengineering, Zhejiang University of Technology, Hangzhou 310014, China
Lysine acetylation is one of the most prevalent and important posttranslational modifications (PTM) with diverse biological functions. In the past years, research findings demonstrate that acetyl-lysine-mediated protein interactions are capable of regulating numerous cellular processes, e.g., chromatin remodeling, DNA repair, cell metabolism and transcriptional activation [1]. For example, lysine acetylation on the histone proteins can neutralize their positive charge to attenuate the electrostatic interaction between the histones and DNAs. The process results in a more expanded chromatin structure that is linked with transcriptionally active genes. Acetylated lysine residues can also serve as the docking sites for reader proteins which could trigger proteinprotein interactions [2].
Bromodomains (BRDs) comprise a family of proteins that selectively interact with acetylated lysine residues (Kac) [3]. BRDcontaining proteins play an important role in regulating various cellular events including gene transcription, cell differentiation and cell cycle control. Malfunction of BRD-containing proteins are reported to be associated with a variety of diseases such as cancer, infectious diseases, diabetes, neurological diseases and others [4]. Many small molecule inhibitors have been developed to inhibit the interactions of acetylated lysine/BRDs. Some of them have become promising lead compounds for treating cancer. For example, two BET bromodomain inhibitors, JQ-1 and I-BET, were found to display effective anti-tumor activities against cancers such as NUT midline carcinoma (NMC) [5].
Inlightof theimportant roles of BRDs in biology, we seektodesign fluorescent probes for detecting BRDs based on the proximityinduced protein conjugation reaction. Biocompatible site-specific covalent protein reactions, which stably tether the target protein with their inhibitors/substrates, greatly facilitate the study of post translational modifications [6], bioimaging of proteins [7] and the creation of protein conjugates for targeted therapeutics [8]. Cysteine mediated reaction is the classical method for proximity-induced protein reaction [9]. Through close examination of the X-ray structure of BRD4(1) [3a], we found that the BRD4(1) protein contains a unique cysteine residue Cys136 at the peptide binding site (Fig. 1), which points toward one of the acetylated lysine residue therefore allows a site-specific covalent probe to be designed (Fig. 1).
|
Download:
|
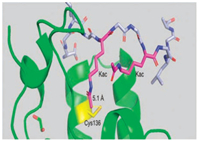
| Fig. 1. Complex structure of BRD4 (1) bound with an acetylated peptide (SGRGKacGGKacGLGY) (PDB id 3uvw). The BRD4(1) domain is shown as cartoon in green, and the peptide ligand shown as ball-and-stick. Cys136 of BRD4(1) domain is highlighted in yellow, and two acetylated lysine residues in magenta. The distance between the sulfur atom of the thiol group and the methyl carbon atom of the acetyl group of acetylated lysine residue (underlined) is measured to be 5.1 Å. | |
As shown in Fig. 2, the probe consists of three parts: 1) a fluorophore that can detect BRD4 activity, 2) a recognition moiety that allows binding to the target protein, and 3) a cysteine reactive site that can form a covalent bond with the target protein. In our design, an environment-sensitive fluorophore NBD was chosen as the reporting unit. Environment-sensitive fluorophores can be highly sensitive to their surrounding environment [10]. The environment-sensitive dye NBD is only weakly fluorescent in water but it emits strong fluorescence in the visible range when transferred to a hydrophobic environment [11]. We envision protein-peptide interaction will bring NBD moiety into a hydrophobic environment of the protein, resulting in the increase of fluorescence emission. It therefore provides a convenient method for in situ monitoring of the protein-peptide interaction.

|
Download:
|
| Fig. 2. Scheme of our probe design. The probe will interact with bromodomain and form a covalent bond through Michael addition reaction. Probe 1 contains an acrylic amide moiety. Probe 2 contains a chloroacetyl group. | |
As reported in the previous research, the peptide SGRGKGGKacGLGY can bind to BRD4(1) efficiently [3a]. In this study we design the probes based on this peptide sequence. A total of two probes were designed with different cysteine-reactive groups to explore which functional group reacts better with BRD4 (1). The two probes are NBD-SGRGKacrylicGGKacGLGY (probe 1) and SGRGKchloroGGKacGLGY (probe 2). The two cysteine-reactive groups are acrylic amide moiety and chloroacetyl group, respectively (Fig. 2). To synthesize the probe, the peptide fragment was first assembled by solid-phase synthesis (Fig. S1 in Supporting information). Fmoc-Lys(Mtt)-COOH was introduced to covalently link the cysteine-reactive group. After the peptide assembly, the NBD fluorophore can be coupled to the N-terminus of the peptide. Subsequently the Mtt group of Fmoc-Lys(Mtt)-COOH was deprotected by diluting TFA and coupled with the two cysteinereactive groups respectively. Finally, the peptide can be cleaved from the resin using TFA. The detail information was deposited in Supporting information.
The two peptide-based probes were first purified by HPLC and characterized by LC–MS (Figs. S2 and S3 in Supporting information). After confirming the purity and the identity of the peptide, we next carried out fluorescence study with BRD4(1). BRD4(1) protein was expressed and purified following the previous protocol [3]. After obtaining the pure protein, BRD4(1) protein was mixed with probes in PBS buffer and incubated for different time points. The fluorescence emission spectra were measured from 500 nm to 650 nm. As shown in Fig. 3A, the result shows that the fluorescence intensity of probe 1 increases with increasing time. It indicates that BRD4(1) can bind to the acrylic peptide efficiently and position the NBD of the probe in a hydrophobic environment, thus leading to the increase of fluorescence. However, in the case of the chloroacetyl-containing peptides, the result shows no obvious change in the fluorescence intensity of probe 2, suggesting that the binding between the chloroacetyl-containing peptide and BRD4(1) protein is not strong enough to induce fluorescence increment (Figs. 3C and D).

|
Download:
|
| Fig. 3. Fluorescence measurement of probes incubated with BRD4(1) in PBS buffer. (A) and (B), time dependent experiment of probe 1 with BRD4(1); (C) and (D), time dependent experiment of probe 2 with BRD4(1). Ex: 460 nm. | |
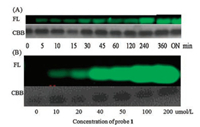
In light of the above results, we chose probe 1 for subsequent protein labelling experiments. BRD4(1) was incubated with probe 1 in PBS buffer for different time points and then analyzed by SDS page experiments. As shown in Fig. 4, a distinct fluorescent band could be observed when BRD4(1) was incubated with probe 1 for 5 min. The fluorescence intensity of BRD4(1) increased with increasing time. The fluorescence increase continued for 6 h. Thereafter, the fluorescence signal plateaued, indicating that the reaction has completed (Fig. 4A). We also carried out protein labelling experiments with probe 2. Under the same conditions, no fluorescent protein band was observed, suggesting that there is no covalent reaction between BRD4(1) and probe 2. We reckon the reason could be the low reactivity of chloroacetyl moiety compared with that of acrylic amide group. Concentration dependent experiments of protein labelling were subsequently performed. Different concentrations of probe 1 were prepared in 10 μmol/L PBS at pH 7.4. They were then mixed with 10 μmol/L BRD4(1) and analyzed by SDS page method. Results showed that with the increasing concentrations of the peptide probe, the NBD fluorescence signal also increases until the concentration reaches 100 μmol/L (Fig. 4B).
|
Download:
|
| Fig. 4. In-gel labelling experiments of probe 1 with BRD4(1). (A) Time-dependent experiment of probe 1 with BRD4(1). (B) Concentration-dependent experiment of probe 1 with BRD4(1). These experiments showed an irreversible bond is formed between probe 1 and BRD4(1). FL: in gel fluorescence scanning (NBD channel); CBB: coomassie blue staining; ON: over night. | |
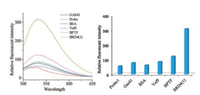
As we know the acetylated peptide can bind to bromodomain proteins in selective manner, we next investigated whether the probe can bind to BRD4(1) specifically. Four other proteins, namely Gas41, BSA, Yaf9 and BPTF, were chosen to incubate with the probe for the selectivity test. In the fluorescence emission experiment, no obvious fluorescence increment was shown by Gas41, BSA and Yaf9. For BPTF protein, slight fluorescence increase was observed (Fig. 5). For the in-gel labelling experiments, only BRD4(1) displayed a strong fluorescent band. Other proteins did not show any fluorescent band (Fig. 6). Taken these results together, probe 1 is proved capable of selectively detecting BRD4(1) and covalently linked to BRD4(1) protein.
|
Download:
|
| Fig. 5. Fluorescence measurement of probe 1 incubated with various proteins in PBS buffer. Ex: 460 nm. Right figure (Em: 535 nm). | |

|
Download:
|
| Fig. 6. In gel labelling experiments of probe 1 with different proteins. FL: in gel fluorescence scanning; CBB: coomassie blue staining. | |
In summary, we have successfully designed and synthesized a fluorescence probe to detect BRD4(1) based on proximity-induced protein conjugation approach. From the result, we can conclude that probe 1, which contains an acrylic amide, shows much stronger interaction with BRD4(1) than probe 2, which contains chloroacetyl group. Probe 1 showed noticeable fluorescence increment when BRD4(1) was added, whereas other proteins did not show obvious fluorescence signal. In-gel labelling experiments proved that a covalent bond is formed between BRD4(1) and probe 1. We envision that the probe developed in this study may provide useful tools for monitoring the activity of BRD4(1) in living cells. Such probe may be useful for screening BRD4(1) inhibitors. We are currently applying this probe to further biological applications.
AcknowledgmentsWe thank Prof. Quan Hao from University of Hong Kong for providing various proteins in this study. We are very grateful for the financial support from the National Natural Science Foundation of China (No. 21572190), the Hong Kong Early Career Scheme Grant (No. 21300714), and the City University of Hong Kong Grant (No. 9667147).
Appendix A. Supplementary dataSupplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.cclet.2018.05.031.
| [1] |
L. Zeng, M.M. Zhou, FEBS Lett. 513 (2002) 124-128. DOI:10.1016/S0014-5793(01)03309-9 |
| [2] |
(a) Y. Y. Li, H. Wen, Y. X. Xi, et al., Cell 159 (2014) 558-571; (b) T. Jenuwein, C. D. Allis, Science 293 (2001) 1074-1080. |
| [3] |
(a) P. Filippakopoulos, S. Picaud, M. Mangos, S. Knapp, et al., Cell 149 (2012) 214-231; (b) D. J. Owen, P. Ornaghi, J. C. Yang, et al., EMBO J. 19 (2000) 6141-6149. |
| [4] |
(a) S. Muller, P. Filippakopoulos, S. Knapp, Mol. Med. 13 (2011) 1-21; (b) T. Nakamura, J. Blechman, S. Tada, et al., Proc. Natl. Acad. Sci. U. S. A. 97 (2000) 7284-7289. |
| [5] |
(a) P. Filippakopoulos, J. Qi, S. Knapp, J. E. Bradne, et al., Nature 468 (2010) 1067-1073; (b) M. A. Dawson, R. K. Prinjha, A. Dittmann, et al., Nature 478 (2011) 529-533. |
| [6] |
(a) C. D. Spicer, B. G. Davis, Nat. Commun. 5 (2014) 4740; (b) Y. S. Yu, J. Xia, Sci. China Chem. 59 (2016) 853-861. |
| [7] |
(a) L. Xue, I. A. Karpenko, J. Hiblot, K. Johnsson, Nat. Chem. Biol. 11 (2015) 917-923; (b) Y. Y. Huang, Y. L. Jin, R. Zhao, Sci. China Chem. 59 (2016) 1250-1257. |
| [8] |
(a) R. V. J. Chari, M. L. Miller, W. C. Widdison, Angew. Chem. Int. Ed. 53 (2014) 3796-3827; (b) V. Chudasama, A. Maruani, S. Caddick, Nat. Chem. 8 (2016) 114-119. |
| [9] |
(a) A. Yang, S. Ha, J. Ahn, et al., Science 354 (2016) 623-626; (b) N. Stephanopoulos, M. B. Francis, Nat. Chem. Biol. 7 (2011) 876-884. |
| [10] |
G.S. Loving, M. Sainlos, B. Trends, Biotechnology 28 (2010) 73-83. |
| [11] |
(a) A. Chattopadhyay, S. Mukherjee, H. Raghuraman, J. Phys. Chem. B 106 (2002) 13002-13009; (b) A. Chattopadhyay, E. London, Biochim. Biophys. Acta 938 (1988) 24-34; (c) S. Fery-Forgues, J. P. Fayet, A. Lopez, J. Photochem. Photobiol. A 70 (1993) 229-243; (d) S. Mukherjee, A. Chattopadhyay, A. Samanta, T. Soujanya, J. Phys. Chem. 98 (1994) 2809-2812. |
 2018, Vol. 29
2018, Vol. 29 


