b University of Chinese Academy of Sciences, Beijing 100101, China;
c State Key Laboratory of Transducer Technology, Chinese Academy of Sciences, Beijing 100101, China
In recent decades, the notable expansion of peptide therapeutics development led to an unprecedented number of marketing approvals and provided a robust pipeline for innovative applications in the near future [1]. As one of the most important strategies in this field, modification of peptides could improve their pharmacological properties, biological efficacies and physicochemical properties [2-12]. Opioid peptides that served as classical peptides have attracted keen attention as promising pharmaceutical agents for pain alleviation owing to their elevated potency and centrally mediated actions of pain process [13-16]. However, their low protease-resistance, inefficiency to penetrate the blood-brain barrier (BBB) and toxic effects severely impede their therapeutic applications [17, 18]. Peptide modification has emerged as an invaluable tool to improve peptide stability and enhance immunogenicity, but there remains a need for additional and complementary reactions with improved kinetics and selectivities [19, 20]. In particular, opioid peptides such as endomorphin-1 (EM1) and endomorphin-2 (EM2) have a biologically indispensable N-terminal tripeptide fragment, and the modification of this fragment would usually influence their affinity to opioid receptors [21]. Therefore, generally applicable methods for highly chemoand regioselective C-terminal functionalization are still urgently demanded [22-24].
C-terminal modification of opioid peptides holds great promise to ameliorate their stability and BBB permeability [25, 26]. More importantly, it could further facilitate the introduction of functional groups, such as fluorescent reporters or synthetic polymers, allowing peptide biophysical studies and applications [27]. Recently, we demonstrated that the computationally engineered (framework for rapid enzyme stabilization by computation, FRESCO strategy) peptide amidase (PAM) could be wellapplied as a versatile catalyst for diverse C-terminal peptide modification reactions [28]. By decreasing the steric hindrance at residue 171, PAM 12B was obtained with enhanced catalytic performance to tolerate bulky nucleophiles such as benzylamine. Herein, we would like to report the application of PAM 12B, which exhibited exciting potentials in the C-terminal modification of opioid peptides by employing prop-2-yn-1-amine (PYA) or prop-2- en-1-amine (PEA) as the nucleophile. A wide range of opioid peptides could be readily functionalized at the C-terminus in a mild and selective fashion (Scheme 1).
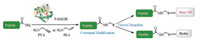
|
Download:
|
| Scheme 1. PAM 12B-catalyzed peptide C-terminal click-functionalization. | |
Initially, we set out to evaluate the catalytic efficiency of PAM 12B in the aminolysis of amide between a commercially available model peptide Cbz-G-Y-NH2 and PYA in organic solvents. Such reaction will provide a platform for the installation of functional groups into the target peptide. As shown in Table 1, in the presence of 9.0 equiv. of PYA, the reaction proceeded efficiently to afford the desired product a1 in 99% yield in 5 h, and only trace amount of hydrolytic product was detected (Table 1, entry 1). Reduction of PYA loading was proven to be less efficient, and longer reaction time was needed to reach the same yield (Table 1, entry 2). Meanwhile, increasing the concentration of PYA to 90 mmol/L had no obvious effect on the reaction efficiency, which indicated saturation kinetics with respect to the nucleophile (Table 1, entry 3). The use of the C-terminal non-protected peptide Cbz-G-Y-OH as the acyl donor was also feasible, and the corresponding product was produced in 99% yield in 6 h (Table 1, entry 4). In the case of PEA as the nucleophile, the reaction somehow turned out to be sluggish, but still afforded the desired product a2 in excellent yield in 12 h (Table 1, entry 5).
|
|
Table 1 Optimization for PAM 12B-catalyzed aminolysis. |
We then sought to explore the modification of opioid peptides (Scheme 2). Enkephalins are one of the most important groups of endogenous opioid peptides [29, 30], which bind to opioid peptide receptors (OPr), including μ opioid peptide receptor (MOPr) and δ opioid peptide receptor (DOPr) [31], and play significant roles in emotions, attachment behaviors, and responses to stress and pain [32, 33]. Since enkephalins share a common 'opioid motif' constituted by the amino acid sequence YGGF, we chose Cbz-YGGFL-NH2 as the prototypical substrate. The reaction of Cbz-YGGFL-NH2 with PYA in the presence of PAM 12B can deliver the corresponding products a3 in 96% yield (Scheme 2). To investigate the practical applicability of PAM 12B-catalyzed peptide C-terminal alkynylation, we also performed this reaction on a 35 mg semi-preparative scale. After work-up and purification the desired product was collected in 70% isolated yield (Scheme S1 in Supporting information).
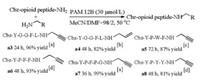
|
Download:
|
| Scheme 2. PAM 12B-catalyzed peptide C-terminal aminolysis. Yields were determined by HPLC. Reaction conditions: [a] peptide (5 mmol/L), PYA (45 mmol/L). [b] peptide (5 mmol/L), PEA (45 mmol/L). [c] peptide (1 mmol/L), PYA (9 mmol/L). [d] peptide (2 mmol/L), PYA (18 mmol/L). | |
Besides PYA, PEA was also a suitable nucleophile for the modification of opioid peptides. As displayed in Scheme 2, CbzYGGFL-NH2 could be well applied in the reaction to give the corresponding products a4 in 82% yield. Encouraged by the initial success, we moved forward to examine the applicability of this approach for miscellaneous opioid peptides. Endomorphins (EMs) were next examined, which are widely distributed in the central nervous system [34, 35]. With Cbz-YPWF-NH2 (EM1) and CbzYPFF-NH2 (EM2) as the substrates, both reactions proceeded well and expected products a5 and a6 were obtained with 87% yield and 93% yield, respectively. Besides the endogenous opioids, exogenous opioids, such as casomorphins, may be released during the digestion of proteins by proteolytic enzymes [36]. In this context, we explored the use of PAM 12B for the modification of Cbz-YPFPG-NH2 (β-casomorphin). This peptide was efficiently converted to the corresponding aminolytic product a7 in 99% yield. Furthermore, we investigated the feasibility to modify the antagonist of opioid receptors. The employment of Cbz-YPYY-NH2 (casoxin B [37]) in this reaction directly afforded the product a8 in 81% yield. This chemistry thus provides a straightforward and novel access to incorporating terminal alkyne and alkene groups into opioid peptides, implying broad applications in their downstream transformations.
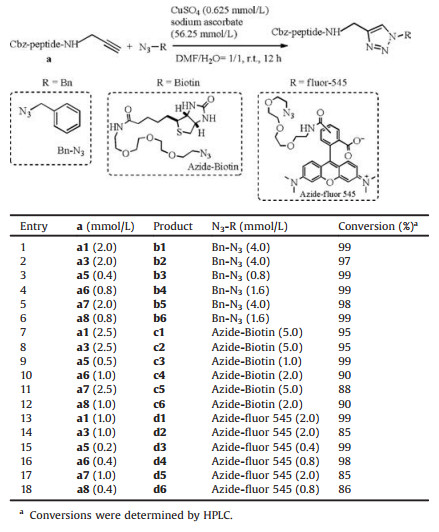
Next, we examined the viability of the opioid peptides functionalization via bioorthogonal chemistry, which plays a significant role in chemical biology research by creating the means to carry out selective chemical transformations in complex biological samples. The aminolytic products were subjected to copper(Ⅰ)-catalyzed azide-alkyne cycloaddition reaction (CuAAC reaction). Initially, benzyl azide, an inexpensive and model azide reagent, was chosen to optimize the reaction conditions. Cbz-G-YNH2 reacted with PYA catalyzed by PAM 12B, then MeCN and residual PYA were removed by heating in boiling water. The unpurified modified peptides were redissolved in DMF and directly subjected to the following CuAAC reaction with benzyl azide as the substrate. The corresponding azole product b1 was obtained in 99% yield in 12 h. Other modified opioid peptides were subsequently produced in 97%-99% yield (Table 2, b2–b6). These results indicated that opioid peptides modification and CuAAC reaction procedures could be carried out consecutively without elaborate isolation of the intermediate, thereby constituting a general, safe and simple method for the functionalization of opioid peptides.
|
|
Table 2 Click reaction of opioid peptides and benzyl azide/azide-biotin/azide-fluor 545. |
To demonstrate the potential biological and medical utilities, azide-biotin and azide-fluor 545 were explored to label opioid peptides. Biotin has been widely used in the fields of catalysis, chemical biology, cell biology, proteomics, detection, labeling and drug delivery for its highly selective and stable interaction with avidin [38]. Bioconjugation of biologically active molecules (like opioid peptides) and biotin-tagged azide is an important method in biotin-avidin system [39]. To date, various opioid peptides can be successfully converted into the corresponding products in excellent yields (88%-99%) (Table 2, c1–c6). To facilitate the molecular mechanistic studies of proteins and medicines, single-molecule fluorescent labeling for attaching fluorophore to specific bioactive molecules has become a pivotal chemical biology technology [40, 41]. Particularly, azide-fluor 545 has been conjugated to proteins as a well-documented model azide with an excitation/ emission peak at 546/565 nm [42, 43]. In this context, we attempted to apply this fluorogenic azide in the click reaction. Remarkably, the results exhibited 85%-99% yield of the corresponding products for diverse types of opioid peptides (Table 2, d1–d6).
In conclusion, with the aid of computational tools, we have developed a robust biocatalyst offering a mild, efficient and selective method for the C-terminal click-functionalization of peptides. Various opioid peptides, along with prop-2-yn-1-amine (PYA) or prop-2-en-1-amine (PEA) as the nucleophile, were smoothly transformed to the corresponding modified opioid peptides with excellent yield and chemo-selectivity. To demonstrate the potential biological and medical utilities, modified peptides bearing alkynyl moieties have been successfully applied in the click reaction with azide-biotin or azide-fluor 545 as the reaction partner. We anticipate that the development of this versatile protocol combined with the power of computational tools will guide the future design and application of the C-terminal functionalization of peptides.
AcknowledgmentsThis work is supported by the National Natural Science Foundation of China (No. 31601412), the 100 Talent Program grant and Biological Resources Service Network Initiative (No. ZSYS-012) and grant from the Chinese Academy of Sciences (No. SKT1604).
Appendix A. Supplementary dataSupplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.cclet.2018.03.033.
| [1] |
A. Kaspar, J.M. Reichert, Drug Discov. Today 18 (2013) 807-817. DOI:10.1016/j.drudis.2013.05.011 |
| [2] |
W.X. Liu, R. Wang, Med. Res. Rev. 32 (2012) 536-580. DOI:10.1002/med.2012.32.issue-3 |
| [3] |
E.M. Sletten, C.R. Bertozzi, Angew. Chem. Int. Ed. 48 (2009) 6974-6998. DOI:10.1002/anie.v48:38 |
| [4] |
H. Noda, G. ErÅs, J.W. Bode, J. Am. Chem. Soc. 136 (2014) 5611-5614. DOI:10.1021/ja5018442 |
| [5] |
M.E.B. Smith, F.F. Schumacher, C.P. Ryan, et al., J. Am. Chem. Soc. 132 (2010) 1960-1965. DOI:10.1021/ja908610s |
| [6] |
A. Dondoni, Angew. Chem. Int. Ed. 47 (2008) 8995-8997. DOI:10.1002/anie.v47:47 |
| [7] |
Y.M. Li, Y.T. Li, M. Pan, et al., Angew. Chem. Int. Ed. 53 (2014) 2198-2202. DOI:10.1002/anie.201310010 |
| [8] |
O. Boutureira, G.J.L. Bernardes, Chem. Rev. 115 (2015) 2174-2195. DOI:10.1021/cr500399p |
| [9] |
L. Schmohl, D. Schwarzer, Curr. Opin. Chem. Biol. 22 (2014) 122-128. DOI:10.1016/j.cbpa.2014.09.020 |
| [10] |
Z. Wu, X. Guo, Z. Guo, Chem. Commun. 46 (2010) 5773-5774. DOI:10.1039/c0cc00828a |
| [11] |
T. Sijbrandij, N. Cukkemane, K. Nazmi, E.C.I. Veerman, F.J. Bikker, Bioconjugate Chem. 24 (2013) 828-831. DOI:10.1021/bc4000146 |
| [12] |
X.L. Tan, M. Pan, Y. Zheng, et al., Chem. Sci. 8 (2017) 6881-6887. DOI:10.1039/C7SC02937C |
| [13] |
M.P. Davis, Expert Opin. Drug Discov. 5 (2010) 1007-1022. DOI:10.1517/17460441.2010.511473 |
| [14] |
B.L. Kieffer, Trends Pharmacol. Sci. 20 (1999) 19-26. DOI:10.1016/S0165-6147(98)01279-6 |
| [15] |
Z.H. Gu, B. Wang, Z.Z. Kou, et al., Neurosignals 25 (2017) 98-116. DOI:10.1159/000484909 |
| [16] |
S. Martin-Schild, J.E. Zadina, A.A. Gerall, S. Vigh, A.J. Kastin, Peptides 18 (1997) 1641-1649. DOI:10.1016/S0196-9781(97)00320-3 |
| [17] |
C. Tömböly, A. Péter, G. Tóth, Peptides 23 (2002) 1573-1580. DOI:10.1016/S0196-9781(02)00100-6 |
| [18] |
A. Janecka, R. Perlikowska, K. Gach, A. Wyrebska, J. Fichna, Curr. Pharm. Design 16 (2010) 1126-1135. DOI:10.2174/138161210790963869 |
| [19] |
C.P. Guimaraes, M.D. Witte, C.S. Theile, et al., Nat. Protoc. 8 (2013) 1787-1799. DOI:10.1038/nprot.2013.101 |
| [20] |
A. El Dahshan, S. Weik, J. Rademann, Org. Lett. 9 (2007) 949-952. DOI:10.1021/ol062754+ |
| [21] |
R. Schwyzer, Biochemistry 25 (1986) 6335-6342. DOI:10.1021/bi00368a075 |
| [22] |
P. Cherkupally, G.A. Acosta, S. Ramesh, et al., Amino Acids 46 (2014) 1827-1838. DOI:10.1007/s00726-014-1746-7 |
| [23] |
J.A. Fehrentz, M. Paris, A. Heitz, et al., J. Org. Chem. 62 (1997) 6792-6796. DOI:10.1021/jo962408d |
| [24] |
L. Ju, J.W. Bode, Org. Biomol. Chem. 7 (2009) 2259-2264. DOI:10.1039/b901198f |
| [25] |
A. Lindqvist, S. Jönsson, M. Hammarlund-Udenaes, Mol. Pharm. 13 (2016) 1258-1266. DOI:10.1021/acs.molpharmaceut.5b00835 |
| [26] |
Y. Koda, M.D. Borgo, S.T. Wessling, et al., Bioorg. Med. Chem. 16 (2008) 6286-6296. DOI:10.1016/j.bmc.2008.04.020 |
| [27] |
G. Zhang, S. Zheng, H. Liu, P.R. Chen, Chem. Soc. Rev. 44 (2015) 3405-3417. DOI:10.1039/C4CS00393D |
| [28] |
B. Wu, H.J. Wijma, L. Song, et al., ACS Catal. 6 (2016) 5405-5414. DOI:10.1021/acscatal.6b01062 |
| [29] |
J. Hughes, T.W. Smith, H.W. Kosterlitz, et al., Nature 258 (1975) 577-579. DOI:10.1038/258577a0 |
| [30] |
S. Udenfriend, D.L. Kilpatrick, Arch. Biochem. Biophys. 221 (1983) 309-323. DOI:10.1016/0003-9861(83)90149-2 |
| [31] |
Y. Li, M.R. Lefever, D. Muthu, et al., Future Med. Chem. 4 (2012) 205-226. DOI:10.4155/fmc.11.195 |
| [32] |
M.S. Henry, L. Gendron, M.E. Tremblay, G. Drolet, Neural. Plast (2017), Article ID 1546125. |
| [33] |
D. Owczarek, D. Cibor, T. Mach, et al., Adv. Med. Sci. 56 (2011) 158-164. DOI:10.2478/v10039-011-0051-x |
| [34] |
J.E. Zadina, L. Hackler, L.J. Ge, A.J. Kastin, Nature 386 (1997) 499-502. DOI:10.1038/386499a0 |
| [35] |
S. Martin-Schild, A.A. Gerall, A.J. Kastin, J.E. Zadina, Peptides 19 (1998) 1783-1789. DOI:10.1016/S0196-9781(98)00136-3 |
| [36] |
D.D. Nguyen, S.K. Johnson, F. Busetti, V.A. Solah, Crit. Rev. Food Sci. Nutr. 55 (2015) 1955-1967. DOI:10.1080/10408398.2012.740102 |
| [37] |
H. Teschemacher, G. Koch, V. Brantl, Biopolymers 43 (1997) 99-117. |
| [38] |
C.M. Dundas, D. Demonte, S. Park, Appl. Microbiol. Biotechnol. 97 (2013) 9343-9353. DOI:10.1007/s00253-013-5232-z |
| [39] |
S. Li, L. Wang, F. Yu, et al., Chem. Sci. 8 (2017) 2107-2114. DOI:10.1039/C6SC02297A |
| [40] |
L. Li, Z. Zhang, Molecules 21 (2016) 1393-1415. DOI:10.3390/molecules21101393 |
| [41] |
B.L. Oliveira, Z. Guo, G.J.L. Bernardes, Chem. Soc. Rev. 46 (2017) 4895-4950. DOI:10.1039/C7CS00184C |
| [42] |
A. Borrmann, S. Milles, T. Plass, et al., Chembiochem 13 (2012) 2094-2099. DOI:10.1002/cbic.v13.14 |
| [43] |
T. Smyth, K. Petrova, N.M. Payton, et al., Bioconjugate Chem. 25 (2014) 1777-1784. DOI:10.1021/bc500291r |
 2018, Vol. 29
2018, Vol. 29