Ras proteins function as molecular switches in some important cellular signaling pathways through cycling between active GTPbound state and inactive GDP-bound state, which regulate plenty of cell processes, including cell survival, proliferation, differentiation, apoptosis and cytoskeletal dynamics [1]. The genetic mutations of RAS genes that persistently activate downstream signaling pathways, can cause cancers. About 30% of human cancers are associated with RAS mutations [2]. In order to trigger the downstream signaling, Ras proteins must bind to the inner face of plasma membrane, which requires the lipid and methyl modifications at the C-terminal tail of proteins. These protein post-translational modifications (PTMs) play a key role in regulating membrane association and cellular process of Ras proteins, like trafficking process and protein-protein interactions [3]. However, lacking appropriate protein tools with PTMs had been a big hindrance to the biofunctional studies of Ras proteins. In recent years, the chemical biology approaches, like protein chemical synthesis and semi-synthesis, have experienced huge advances and brought great chances to obtain Ras proteins with multi-modifications. These synthesized Ras proteins largely facilitate the related functional research. Thus, this mini-review mainly summarizes recent strategies for the synthesis of Ras proteins and their applications in biofunctional studies.
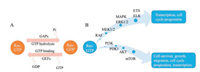
2. Ras proteinsRas proteins are guanine nucleotide-bound proteins (G proteins), which are encoded by RAS genes. RAS subfamily contains three genes (NRAS, HRAS and KRAS), which can translate into four protein isoforms: N-Ras, H-Ras, K-Ras4A and K-Ras4B [4]. All of Ras proteins are composed of two parts: one is the G domain (the first 165 amino acids), which is highly conserved; the other is the hypervariable region (HVR), which contains approximately 20 amino acids (Fig. 1). Ras proteins are under a dynamic equilibrium between 'on' and 'off' conformations which are conferred by GTP and GDP exchanging. Normally, the active GTP-bound state can be switched to the inactive GDP-bound state by GTPase activating proteins (GAPs, accelerating GTP hydrolysis and GDP binding). The exchange of GDP-Ras for GTP-Ras is regulated by guanine nucleotide exchange factors (GEFs, stimulating the release of GDP) [1].

|
Download:
|
| Fig. 1. The hypervariable region (HVR) of four Ras proteins. N-Ras and K-Ras4A contains palmitoyl and farnesyl groups. H-Ras contains two palmitoyl and farnesyl groups. KRas4B contains farnesyl group and polybasic lysine. Reproduced with permission [5]. Copyright 2014, Elsevier Ltd | |
Ras proteins play a significant role in cellular signaling pathways and regulate a serial of biological events, including cell survival, cell adhesion, apoptosis and cellular metabolism. When Ras proteins bind to GTP, they trigger the downstream signals. There are two main Ras signaling pathways, MAPK (RAF/MEK/ERK) pathway and PI3K/PDK/AKT pathway [5]. The MAPK pathway requires the dimerization of Ras. Dimerized Ras activates RAF, which promotes the phosphorylation of MEK1/2, and further activates the ERK1/2, resulting in the growth of cell. Ras can also effectively activate PI3K/PDK/AKT pathway. Receptor tyrosine kinase (RTK) and Ras binding to PI3K give rise to the phosphorylation of PI3K, which recruit the AKT with the help of PDK1, leading to the activation of mTOR. (Fig. 2B) [6].
|
Download:
|
| Fig. 2. The cycle and signaling pathways of Ras proteins. (A). GTP-Ras is the active form, and the GTP can be hydrolyzed with the help of GAPs, resulting in the inactive GDPbound Ras; GDP-Ras is the inactive form, and the GDP can be released with the help of GEFs, resulting in the active GTP-bound Ras. (B). Two main Ras-involved signaling pathways in cellular process. One is MAPK pathway, another one is PI3K pathway. Reproduced with permission [5]. Copyright 2014, Elsevier Ltd. | |
In order to activate the downstream signaling, Ras proteins must localize to the plasma membrane, and the PTMs of Ras proteins are indispensable for their enrichment on the plasma membrane. The nascent Ras proteins contain a CAAX motif at its C-terminus, which is further modified by a series of enzymes, including prenyltransferases (farnesyltransferase or geranylgeranyltransferase type Ⅰ, adding farnesyl or polyisoprene group to the CAAX Cys), RAS-converting enzyme 1 (RCE1, removing the AAX motif), isoprenyl cysteine transferase (Icmt, adding the methyl ester to the carboxyl group of farnesylcysteine) [7]. These modifications largely enhance the binding affinity of Ras proteins to the inner face of plasma membrane for triggering the downstream signals (Fig. 3A).

|
Download:
|
| Fig. 3. The process of Ras proteins' PTMs and the frequency of Ras mutations in various human cancers. (A) Ras proteins undergo three enzymes-catalyzed reactions at least to get the lipid and methyl modifications for targeting the plasma membrane [4]. (B) The mutation frequencies of Ras proteins differ in disparate cancers [9] | |
The activated RAS genes due to mutation were identified in human cancer for the first time in 1982 [8, 9]. Subsequent research found that RAS genes are among the most frequently mutated genes in human cancer. 97% of pancreatic ductal adenocarcinoma and 52% of colorectal adenocarcinoma are related to KRAS mutations, 29% of skin cutaneous melanoma and 13% of thyroid carcinoma are associated with NRAS mutations, and 11% of Bladder urothelial carcinoma are relevant to HRAS mutations [6, 9] (Fig. 3B). Thus, Ras proteins are important anticancer drug targets, and scientists have dug into them for many years. However, decades of research have not yet produce a Ras-targeting drug for clinical use. In the early days, the research focused on interfering GTP binding to the Ras proteins [10]. But Ras proteins turned out to have a high affinity for GTP so that the GDP-like drugs were not effective [11-13]. Later, the research moved on to destroy the function of Ras proteins by inhibiting farnesyltransferase to disturb the binding affinity of Ras proteins and plasma membrane [9, 14-16]. It looked like a winning solution, but unfortunately it failed in clinical trials. Besides, there are no suitable pockets for small molecules to bind with Ras proteins' surfaces. Ras proteins are so called "undruggable" [17].
Thus, uncovering the mechanisms of regulating Ras proteins' activity is important for developing anti-Ras therapeutics.
Over the past decades, scientists have spared no efforts on figuring out the whole structure of Ras proteins, as well as the role of modified Rasproteins in cellularprocesses.Due to the flexibilityand complexity of the C-terminal structure in Ras proteins, these questions have been difficult to be fully addressed. Thus, the scientists made efforts to prepare the full-length Ras proteins with multi-modifications for exploring the function of Ras protein, the mechanism of Ras signaling, and anti-Ras drug design. In this review, we summarize current strategies for obtaining Ras proteins with multi-modifications, and their applications for in vitro and in vivo biofunctional studies.
3. Strategies for obtaining Ras proteinsTill now, there are various approaches to obtain Ras proteins, such as purifying from E. coli cells [18, 19] or particular mammalian tissues [20], chemical total synthesis, and semi-synthesis. In this chapter, we mainly introduce the methods of obtaining Ras proteins with modifications.
3.1. Total synthesis of Ras proteinsWith the advancing of protein synthesis technology, it becomes not so difficult to synthesize desired proteins. Remarkably, Kent and Muir devised simple techniques of native chemical ligation (NCL) and expressed protein ligation (EPL), which indicated that the field of protein synthesis entered a new era [21-23]. After that, more and more innovative protein ligation techniques have been developed, such as serine/threonine ligation [24-28], peptide hydrzides-mediated NCL [29-31]. Meanwhile, many proteins with different PTMs have been obtained through protein chemical synthesis or semi-synthesis, such as phosphorylated proteins, glycosylated proteins, ubiquitinated proteins and lipidated proteins [32-40].
In 2017, Danishefsky's group prepared two truncated K-Ras (G12 V) proteins variant (1–166) containing all-L or all-D type amino acids without PTMs via chemical total synthesis [41]. Although it is easy to obtain the all-L K-Ras through recombinant protein expression in E. coli, the all-D variant can only be produced via chemical synthesis. This protein material enables the mirrorimage display screening to identify all-D peptide inhibitors of oncogenic K-Ras, which facilitates developing anti-Ras therapies. However, the comparative low yield of chemical total synthesis limits the preparative scale of Ras proteins.
3.2. Biosynthesis of Ras proteins with modificationsDue to the importance of the C-terminal PTMs of Ras proteins, Ras proteins with lipidation were acquired through the incubation of nascent protein with farnesyl transferase to study the MAP kinase activation system [42, 43]. However, it cannot fully represent the lipidated Ras proteins in vivo, which undergo several further enzyme-catalyzed modifications. In 2015, Stephen's group obtained full-length K-Ras4B proteins with farnesylation and methylation using the insect cell expression system [44], which is a big breakthrough for the preparation of K-Ras4B proteins.
3.3. Semi-synthesis of Ras protein with modificationsThough recombinant protein expression is a useful method to obtain Ras proteins, there still exist lots of limits. For example, the common prokaryote systems like E. coli lack machines to undergo protein posttranslational modifications; the eukaryote systems often suffer from low protein expression level and heterogeneous PTMs. Chemical total synthesis provides much more freedom to introduce all kinds of modifications into proteins, but it is often expensive and time-consuming. By combining the advantages of both protein expression and chemical synthesis, protein semisynthesis has become a powerful strategy to prepare proteins with homogeneous multi-PTMs in a considerable amount [45, 46]. Ras proteins contain important lipid and methyl modifications in the C-terminal. Thus, according to the protein semi-synthesis strategy, the N-terminal parts of Ras proteins without PTMs can be easily obtained via recombinant protein expression while the C-terminal peptides with PTMs can be produced through chemical synthesis. In the end, both two parts are ligated to generate full-length Ras proteins with multi-PTMs. Moreover, additional tags like fluorophores and isotope elements for biophysical assays can also be site-specifically incorporated into synthesized proteins.
3.3.1. The strategies of lipopeptides synthesisThe HVR regions of Ras proteins often contain more than one PTM including lipids and methyl ester C-terminus. In particular, farnesyl group is acid-labile, which is not compatible with common peptide cleavage and deprotection conditions using strong acids. A variety of methods to synthesize lipopeptide and ligation strategies to incorporate the peptides into Ras proteins have been developed [47]. With the technique of peptide synthesis developed continuously, the strategies of lipopeptides synthesis have shifted from liquid phase synthesis to solid phase synthesis, from Boc protecting group strategies to Fmoc protecting group strategies, from lipidated amino acids building block synthesis to lipidation of peptides site specifically. The yield and simplicity increased significantly and continuously.
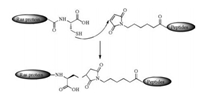
Waldmann's group synthesized S-palmitoylated and S-farnesylated C-terminal peptide of the human N-Ras protein by using dipeptides building blocks, of which acetate protecting group can be deprotected in a mild condition by lipase from Mucor miehei [48] (Scheme 1). The cysteine with farnesyl and methyl modifications were synthesized in classical chemical methods as a starting point and then attached with next dipeptide building blocks in a liquid phase. The combination of classical chemical synthesis methods and enzymatic technique can help to obtain various Ras lipopeptides.
|
Download:
|
| Scheme 1. The dipeptides building blocks synthesis in a mild condition with lipase from Mucor miehei | |
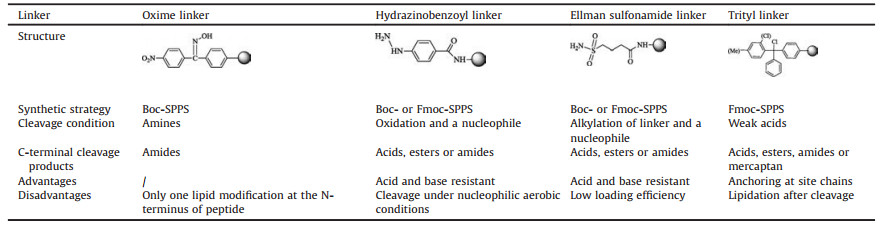
With the linker strategies developed, the pre-lipopetides can be anchored on different kinds of resins, and be further elongated following solid phase peptide synthesis (SPPS) strategies (Table 1) [49]. Thus, the modification sites were first protected with different groups, which were respectively deprotected and further transformed into different PTMs site-specifically. In 2001, Poulter's group [50] first used Kaiser benzophenone oxime linker to synthesize a CAAX region peptide of Ras protein with a farnesyl group at the N-terminal cysteine using Boc-based SPPS strategy. Additional biotin tag was attached on the N-terminus of the tetrapeptide and then it was released under a mild base condition.
|
|
Table 1 Different linkers for solid-phase peptides synthesis of lipopeptides [49] |
Waldmann's group improved the linker strategies so that lipopeptides were easily synthesized using other two kinds of linkers. With an aryl hydrazide linker [51], the resin loading process and elongation of peptide were carried on following Fmocbased SPPS strategy. Hydrazide linker was cleaved under nucleophilic aerobic conditions, and the obtained peptide could be either an ester or an amide. H-Ras and N-Ras peptides with two or three lipids modification and methylation were firstly synthesized by this strategy. Another one is Ellman alkyl sulfonamide linker [52, 53]. This linker is stable to be treated with either acid or base condition. The target peptides were released under a mild condition by selective N-alkylation and attack of different nucleophiles, obtaining a product of an ester or an amide. HRas, N-Ras and K-Ras4B peptides with one or two lipid modification and methylation reached a yield of over 60%.
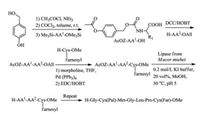
In 2012, Distefano's group synthesized peptides with cysteine methyl esters C-terminus through Trt-Cl resins, which can befurther farnesylated in liquid phase after their releasing from resins [54]. They developed a new strategy for synthesizing peptides containing a C-terminal cysteine methyl ester. According to this strategy, an Fmoc-Cys-OMe building block was first attached onto 2-Cl Trt-Cl or Trt-Cl resins via its side-chain thiol group, and the peptide sequence was further elongated following standard Fmoc-based SPPS procedures. In the end, the free thiol group of the peptide Cterminal cysteine methyl ester was modified by farnesyl group with trans, trans farnesyl bromide after the cleavage of peptides from resins.Otherlipopeptideswithmulti-PTMs, for example C-terminal of LC3-II protein and Rheb protein, can be obtained using the same linker strategies [55-57]. In 2017, our group firstly semi-synthesized the full-length K-Ras4B protein modified with phosphate, farnesyl and methyl groups [58]. The farnesyl moiety cannot accommodate with strong acids, which is applied in the cleavage of phosphoryl ester. Thus, we employed the similar strategy with Distefano's one to add the farnesyl group into the peptide in liquid phase afterits releasingfrom resinsbyTFA. Finally, theK-Ras4BHVR region peptide containing these three modifications was successfully synthesized in a high yield. With a cysteine attached in the N-terminal of the peptide, it was easily coupled with expressed K-Ras4B protein thioester by means of NCL reaction.
3.3.2. The strategies of protein conjugationAfter the synthesis of lipopeptides, the way to attach the peptides to expressed truncated protein is another key factor affecting the yield of protein synthesis.
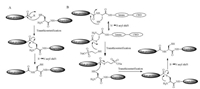
Kent and his coworkers developed NCL reaction in 1994 [21], leading to a high yield of peptide conjugation products (Scheme 2A). The NCL reaction involves two reactants. One is peptide C-terminal thioester and the other is peptide with a cysteine residue at its N-terminus. The thiol group of N-terminal cysteine of one peptide attacks the C-terminal thioester group of another peptide to form a new peptide thioester. Then, the amino group of cysteine attacks the new peptide thioester to undergo an S, N-acyl shift and finally a native amide bond forms between two peptide reactants.
|
Download:
|
| Scheme 2. Two major methods for protein native ligation [21, 22, 47]. (A) Native chemical ligation. (B) Expressed protein ligation | |
Protein thioester can be generated through the split of protein intein strategy and be further used to link with another peptide or protein fragment containing an N-terminal cysteine via NCL [22] (Scheme 2B). This strategy is called expressed protein ligation (EPL). With these powerful techniques, it is possible to obtain Ras protein with multi-PTMs easily. Take a process of Ras protein synthesis as an example, the expressed truncated protein fused to intein can be pulled down according to the designed affinity tag binding to the beads. The purified protein fusion undergoes an S, N-acyl shift and a transthioesterification reaction with MESNa (2- mercaptoethanesulfonate), resulting a newly formed protein thioester. Through an NCL reaction the lipopeptides can be coupled with the protein thioester easily. In 2006, Waldmann's group firstly used an intein mediated EPL reaction for attaching the lipopeptides to expressed protein thioesters and obtained functional H-Ras and K-Ras4B proteins [59]. In 2010, this group also synthesized functional Rheb protein according to the EPL strategy [57].
Besides powerful NCL and EPL, there are other methods to conjugate lipopeptides with the expressed proteins. The MIC ligation reaction (Scheme 3) is an efficient method in protein engineering, which proceeds under a neutral pH condition between a maleimido group within a peptide and a cysteine residue at the C-terminus of truncated protein. The thiol group of cysteine attacks the maleimido group and forms a stable addition product, so that the peptides with multi-PTMs can be incorporated into truncated Ras proteins mildly. In 2000, Waldmann's group synthesized functional H-Ras lipoproteins with fluorescent tags for in vivo studies using MIC ligation [60]. In 2003, they improved the methods and succeeded in synthesizing functional H-Ras and NRas lipoproteins on solid support [61].
Pollok's group reported sortase-mediated ligation as a useful tool for protein engineering in 2004 (Fig. 4) [62]. Sortase A, a kind of transpeptidase, can recognize the specific peptide sequence (Leu-Pro-Glu-Thr-Gly) of substrate and cleave this motif after the Thr residue to form a protein thioester. Then, the Gly residue at the N-terminus of lipopeptide can attack the protein thioester leading to the formation of a native amide bond. In 2012, Dementiev's group used a sortase ligation for obtaining functional K-Ras4B protein with farnesyl and methyl modifications, which was further applied to study K-Ras4B protein's properties and dimer formation [63].

|
Download:
|
| Fig. 4. Sortase-mediated ligation in K-Ras4B protein synthesis. Reproduced with permission [63]. Copyright 2012, Elsevier Inc | |
As we can see, the ligation reactions play a key role in the semisynthesis of Ras protein due to their high yield and simplicity, although they may introduce an insertion of additional cysteine residue or other linkers into Ras protein.
3.4. Summary and prospects of Ras protein synthesisAll these strategies introduced above to obtain Ras proteins with PTMs have their own advantages and disadvantages. Chemical total synthesis is an effective way to obtain Ras proteins with all kinds of PTMs, but it is often expensive and timeconsuming. In addition, this strategy sometimes meets the difficulty of refolding synthesized protein correctly. The biosynthesis approach is potential to generate a great amount of Ras proteins with PTMs in an economic way. However, the produced proteins in eukaryote system usually contain incomplete and heterogeneous PTMs. The semi-synthesis strategy combines the advantages of both chemical synthesis and recombinant protein expression. In particular, the important PTMs of Ras proteins including farnesylation, methylation and phosphorylation mainly localize in their C-terminal region. Thus, the semi-synthesis strategy appears to be the most useful one to generate Ras proteins with multi-PTMs in milligram scale for further application in biofunctional studies.
4. Biophysical researches with synthesized Ras proteinsRas proteins bind to the inner face of plasma membrane with the help of lipidated HVR peptides, and initiate the downstream signaling pathway. Besides, lipidation is also responsible for the subcellular localization of Ras proteins and their interaction with the chaperon protein for transportation [64]. Ras proteins have been studied since 1980 s, while most biochemical studies and structural research of these proteins have been done using truncated proteins without the C-terminal HVR or full-length proteins produced in bacteria without PTMs. For the purpose of figuring out the structure and function of Ras proteins with PTMs in vitro and in vivo, it is essential to obtain the multi-modified proteins in a high yield and quality [44]. With the development of efficient synthetic strategies for generating Ras proteins with multi-modifications, more and more biofunctional research employing these material have been carried out and addressed a serial of important issues that could not be answered in previous time. In this part, we mainly focus on the biophysical studies using synthesized Ras proteins.
4.1. Ras proteins in the membraneThe association with plasma membrane is necessary for Ras proteins to transduce the downstream signal. But the molecular mechanism of Ras proteins-plasma membrane interaction had been not understood in depth before the multi-modified Ras proteins, especially the one with prenylation, were obtained. Scientists have done a lot of cellular research by the means of overexpressing Ras proteins, knocking out the genes of relative enzymes and other in vivo technique. Nevertheless, there still existed a lot of questions about the interaction between Ras proteins and plasma membrane that could not be well addressed through the in vivo studies. For example, how Ras proteins interact with plasma membrane at molecular level, how Ras proteins change their conformation in plasma membrane, and in which subdomain of plasma membrane Ras proteins localize. However, the biofunctional research using synthetic Ras proteins with lipid modification provided a lot of information for answering these questions [65].
The research on lipidated Ras proteins was promoted by the development of protein semi-synthesis strategies. In 1990 s, Gelb's group obtained H-Ras protein with farnesyl modification incorporated by use of farnesyltransferase, and the following research showed that farnesylation is critical for the Ras signaling [42, 43]. Kuhlmann and his co-workers obtained the H-Ras protein containing lipidated HVR by use of semi-synthesis strategy for the first time, which was further proved to be a valuable tool for biophysical researches [66].
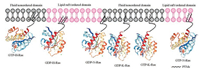
However, it is difficult to carry out the experiments between lipidated proteins and the plasma membrane since the phospholipid in the membrane is similar to the lipidated chains of proteins, which makes the lipid chains and membrane component undistinguishable. To tackle this problem, a lot of new method and technology had been proposed, like fluorescence probes, 2H-labelling of the lipid chains [67-71]. Gorfe's group made use of the triply lipidated peptides of H-Ras to study the behavior of Ras proteins in different membrane domain. The data showed that lipidated chains play an important role in H-Rasmembrane association as well as forming protein nanoclustering in different membrane nanodomains [72]. Both Winter's and Huster's groups took the advantage of semi-synthetic lipidated proteins for a serial biophysical research including investigating the chain length of proteins adapted to the membrane and studying the distribution of proteins in multiphasic artificial membrane, [73-75]. Their research came to conclusions that HRas and N-Ras were enriched in liquid order domains of membrane, whereas K-Ras4B stayed in liquid disorder domains of membrane (Fig. 5).
|
Download:
|
| Fig. 5. The specific affinity of different Ras isoforms with different microdomains of plasma membrane. Reproduced with permission [76]. Copyright 2015, American Association for Cancer Research | |
4.2. Ras proteins in the transport system
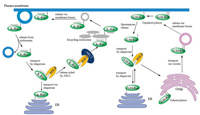
In order to target plasma membrane, N-Ras and H-Ras were transported by vesicle from Golgi through secretory pathway, after they were palmitoylated in the Golgi (Fig. 6) [77]. As for K-Ras4B, it does not contain additional cysteine in its HVR for palmitoylation. Thus, it was transported via an uncharacterized pathway [78]. Some of shuttle proteins were proposed in this trafficking pathway [79, 80], and the most promising one is GDI-like solubilizing factor PDEδ. After Waldmann's group obtained modified Rheb and KRas4B proteins via semi-synthesis [57], Wittinghofer's group further reported the complex structure of PDEδ and farnesylated Rheb, which provided a great explanation to the mechanism of these two proteins' interaction [81]. Meanwhile, Bastiaens's group did a serial of experiments in vivo, demonstrating that PDEδ could sequester farnesylated K-Ras4B in accord with Wittinghofer's data in vitro and regulate the cellular localization of K-Ras4B proteins [82]. Recently, Simanshu's group obtained the multi-modified KRas4B proteins using Stephen's insect system [44], and got the complex structure of multi-modified K-Ras4B and PDEδ [83]. They verified those farnesyl and methyl group are essential for the interaction of K-Ras4B with PDEδ.
|
Download:
|
| Fig. 6. The transport system of different Ras isoforms. Reproduced with permission [5]. Copyright 2014, Elsevier Ltd | |
Due to the fluidity of plasma membrane, the depth of isoprenylation inserting into the plasma membrane is alterable, and Ras proteins can release from the plasma membrane, thus regulating cellular signaling [71, 76, 82]. K-Ras4B protein dissociated from the plasma membrane enters into the cytoplasm and interacts with PDEδ. The Arf-like GTPase Arl2 can bind to the allosteric site of PDEδ, leading to the conformational change of PDEδ and release of K-Ras4B from PDEδ [81]. After that, K-Ras4B is in the surrounding area of nucleus where recycling endosome (RE) is rich. K-Ras4B prefers to bind to the RE membrane, because of the negatively charged surface of RE membrane [84].
When H-Ras contains two palmitoyl groups, it displays a high binding affinity to plasma membrane and cannot spontaneously dissociate from it. In the state of depalmitoylation, H-Ras is soluble in the cytoplasm and can interact with PDEδ. N-Ras contains one palmitoyl group and the process of depalmitoylation will decrease its association with the membrane too. Then, N-Ras is delivered to the Golgi by PDEδ (Fig. 6) [85].
In addition, some other factors including protein phosphorylation and Ca2+/calmodulin (CaM) can also affect the association of K-Ras4B with plasma membrane. The cellular K-Ras4B can be phosphorylated at the site of Ser181 [58, 86, 87]. Our group has semi-synthesized K-Ras4B proteins possessing phosphate, farnesyl and methyl moieties, and further employed them to explore the effects of phosphorylation on interaction between K-Ras4B and membrane by means of biophysical techniques like fluorescence polarization, frequency-domain fluorescence anisotropy, and infrared reflection-absorption spectroscopy. Our studies demonstrate that phosphorylation of K-Ras4B at Ser181 decreases its binding affinity with membrane, in a membrane fluidity-dependent manner. Meanwhile, this phosphorylation at Ser181 does not weaken K-Ras4B's binding with its transport protein PDEδ in cytoplasm. Besides, phosphorylated K-Ras4B maintains the localization in the liquid-disorder domain of membrane [58]. It has been found that the poly-lysine domain of K-Ras4B can interact with Ca2+/CaM, and the farnesyl group can bind to the pocket of Ca2+/CaM [88, 89]. Winter's group has done some biophysical research using semi-synthesized K-Ras4B proteins combined different techniques, like Surface plasmon resonance, atomic force microscope, etc. The results proved that Ca2+/CaM can sequester membrane-bound K-Ras4B and the interaction between two proteins is independent of Ras guanine nucleotide state [89].
5. SummaryRas proteins play a key role in some important cellular signaling pathways that regulate many essential cell events. Mutational activation of Ras proteins is among the most powerful drivers of cancer. The nascent Ras proteins undergo a serial of posttranslational modification reactions to possess lipids and methyl moieties, which are required for their association with plasma membrane to trigger the downstream signaling. So, it is very necessary to obtain Ras proteins with all kind of PTMs for investigating their biofunction. The research history of Ras proteins has seen the great advantages of protein synthesis. As we summarized in this review, chemical total synthesis, biosynthesis and semi-synthesis strategies have been developed for obtaining Ras proteins with PTMs. The three strategies have their own advantages and disadvantages. Generally, Ras protein semisynthesis is more popular since it combines the flexibility of chemical synthesis and the efficiency of protein expression. Meanwhile, the synthesized multi-modified Ras proteins provide a great chance to investigate their biological function deeply at molecular level, including the behaviors of Ras proteins in the membrane (like their mobility, orientation and association with the plasma membrane), as well as protein-protein interactions between Ras and their transport proteins.
AcknowledgmentWe are grateful to the National Natural Science Foundation of China (Nos. 21672125, 91753122).
| [1] |
Y. Pylayeva-Gupta, E. Grabocka, D. Bar-Sagi, Nat. Rev. Cancer 11 (2011) 761-774. DOI:10.1038/nrc3106 |
| [2] |
I.A. Prior, P.D. Lewis, C. Mattos, Cancer Res. 72 (2012) 2457-2467. DOI:10.1158/0008-5472.CAN-11-2612 |
| [3] |
G. Triola, H. Waldmann, C. Hedberg, ACS Chem. Biol. 7 (2012) 87-99. DOI:10.1021/cb200460u |
| [4] |
I.M. Ahearn, K. Haigis, D. Bar-Sagi, M.R. Philips, Nat. Rev. Mol. Cell Biol. 13 (2011) 39-51. |
| [5] |
H. van Hattum, H. Waldmann, Chem. Biol. 21 (2014) 1185-1195. DOI:10.1016/j.chembiol.2014.08.001 |
| [6] |
R. Nussinov, C.J. Tsai, H. Jang, Cancer Res. 78 (2018) 593-602. DOI:10.1158/0008-5472.CAN-17-2727 |
| [7] |
J. Omerovic, A.J. Laude, I.A. Prior, Cell Mol. Life Sci. 64 (2007) 2575-2589. DOI:10.1007/s00018-007-7133-8 |
| [8] |
A.D. Cox, C.J. Der, Small GTPases 1 (2010) 2-27. DOI:10.4161/sgtp.1.1.12178 |
| [9] |
A.D. Cox, S.W. Fesik, A.C. Kimmelman, J. Luo, C.J. Der, Nat. Rev. Drug Discov. 13 (2014) 828-851. DOI:10.1038/nrd4389 |
| [10] |
A.T. Baines, D. Xu, C.J. Der, Future Med. Chem. 3 (2011) 1787-1808. DOI:10.4155/fmc.11.121 |
| [11] |
J. John, H. Rensland, I. Schlichting, et al., J. Biol. Chem. 268 (1993) 923-929. |
| [12] |
I. Becher, M.M. Savitski, M.F. Savitski, et al., ACS Chem. Biol. 8 (2013) 599-607. DOI:10.1021/cb3005879 |
| [13] |
P.M. Cromm, J. Spiegel, T.N. Grossmann, H. Waldmann, Angew. Chem. Int. Ed. Engl. 54 (2015) 13516-13537. DOI:10.1002/anie.201504357 |
| [14] |
J.M. Bergman, M.T. Abrams, J.P. Davide, et al., Bioorg. Med. Chem. Lett. 11 (2001) 1411-1415. DOI:10.1016/S0960-894X(01)00240-2 |
| [15] |
N.M. Appels, J.H. Beijnen, J.H. Schellens, Oncologist 10 (2005) 565-578. DOI:10.1634/theoncologist.10-8-565 |
| [16] |
A.D. Cox, C.J. Der, M.R. Philips, Clin. Cancer Res. 21 (2015) 1819-1827. DOI:10.1158/1078-0432.CCR-14-3214 |
| [17] |
H. Ledford, Nature 520 (2015) 278-280. DOI:10.1038/520278a |
| [18] |
M. Gross, R.W. Sweet, G. Sathe, et al., Mol. Cell Biol. 5 (1985) 1015-1024. DOI:10.1128/MCB.5.5.1015 |
| [19] |
P.N. Lowe, M.J. Page, S. Bradley, et al., J. Biol. Chem. 266 (1991) 1672-1678. |
| [20] |
T. Yamashita, K. Yamamoto, et al., J. Biol. Chem. 263 (1988) 17181-17188. |
| [21] |
P.E. Dawson, T.W. Muir, I. Clark-Lewis, S.B. Kent, Science 266 (1994) 776-779. DOI:10.1126/science.7973629 |
| [22] |
T.W. Muir, D. Sondhi, P.A. Cole, Proc. Natl. Acad. Sci. U. S. A. 95 (1998) 6705-6710. DOI:10.1073/pnas.95.12.6705 |
| [23] |
H. Li, S. Dong, Sci. China Chem. 60 (2016) 201-213. |
| [24] |
X. Li, H.Y. Lam, Y. Zhang, C.K. Chan, Org. Lett. 12 (2010) 1724-1727. DOI:10.1021/ol1003109 |
| [25] |
Y. Zhang, C. Xu, H.Y. Lam, C.L. Lee, X. Li, Proc. Natl. Acad. Sci. U. S. A. 110 (2013) 6657-6662. DOI:10.1073/pnas.1221012110 |
| [26] |
C.L. Lee, H. Liu, C.T. Wong, H.Y. Chow, X. Li, J. Am. Chem. Soc. 138 (2016) 10477-10484. DOI:10.1021/jacs.6b04238 |
| [27] |
C.L. Lee, X. Li, Sci. China Chem. 59 (2016) 1061-1064. |
| [28] |
C. Xu, J. Xu, H. Liu, X. Li, Chin. Chem. Lett. 29 (2018) 1119-1122. DOI:10.1016/j.cclet.2018.03.012 |
| [29] |
G.M. Fang, Y.M. Li, F. Shen, et al., Angew. Chem. Int. Ed. Engl. 50 (2011) 7645-7649. DOI:10.1002/anie.201100996 |
| [30] |
G.M. Fang, J.X. Wang, L. Liu, Angew. Chem. Int. Ed. Engl. 51 (2012) 10347-10350. DOI:10.1002/anie.201203843 |
| [31] |
J.S. Zheng, S. Tang, Y.K. Qi, Z.P. Wang, L. Liu, Nat. Protoc. 8 (2013) 2483-2495. DOI:10.1038/nprot.2013.152 |
| [32] |
M.K. Tarrant, H.S. Rho, Z. Xie, et al., Nat. Chem. Biol. 8 (2012) 262-269. DOI:10.1038/nchembio.771 |
| [33] |
N. Yamamoto, Y. Tanabe, R. Okamoto, P.E. Dawson, Y. Kajihara, J. Am. Chem. Soc. 130 (2008) 501-510. DOI:10.1021/ja072543f |
| [34] |
P. Wang, S. Dong, J.A. Brailsford, et al., Angew. Chem. Int. Ed. Engl. 51 (2012) 11576-11584. DOI:10.1002/anie.201206090 |
| [35] |
K.S. Ajish Kumar, M. Haj-Yahya, D. Olschewski, H.A. Lashuel, A. Brik, Angew. Chem. Int. Ed. Engl. 48 (2009) 8090-8094. DOI:10.1002/anie.v48:43 |
| [36] |
S. Tang, L.J. Liang, Y.Y. Si, et al., Angew. Chem. Int. Ed. Engl. 56 (2017) 13333-13337. DOI:10.1002/anie.201708067 |
| [37] |
Y.W. Wu, L.K. Oesterlin, K.T. Tan, et al., Nat. Chem. Biol. 6 (2010) 534-540. DOI:10.1038/nchembio.386 |
| [38] |
F. Li, L. Yi, L. Zhao, et al., Proc. Natl. Acad. Sci. U. S. A. 111 (2014) 2572-2577. DOI:10.1073/pnas.1313655111 |
| [39] |
M. Pan, S. Gao, Y. Zheng, et al., J. Am. Chem. Soc. 138 (2016) 7429-7435. DOI:10.1021/jacs.6b04031 |
| [40] |
Q. He, J. Li, Y. Qi, et al., Sci. China Chem. 60 (2016) 621-627. |
| [41] |
A.M. Levinson, J.H. McGee, A.G. Roberts, et al., J. Am. Chem. Soc. 139 (2017) 7632-7639. DOI:10.1021/jacs.7b02988 |
| [42] |
P. McGeady, S. Kuroda, K. Shimizu, Y. Takai, M.H. Gelb, J. Biol. Chem. 270 (1995) 26347-26351. DOI:10.1074/jbc.270.44.26347 |
| [43] |
T. Dudler, M.H. Gelb, Biochem. 36 (1997) 12434-12441. DOI:10.1021/bi971054x |
| [44] |
W.K. Gillette, D. Esposito, M. Abreu Blanco, et al., Sci. Rep. 5 (2015) 15916. DOI:10.1038/srep15916 |
| [45] |
S. Kent, Bioorg. Med. Chem. 25 (2017) 4926-4937. DOI:10.1016/j.bmc.2017.06.020 |
| [46] |
X. Bi, K.K. Pasunooti, C.F. Liu, Sci. China Chem. 61 (2017) 251-265. |
| [47] |
T. Mejuch, H. Waldmann, Bioconjugate Chem. 27 (2016) 1771-1783. DOI:10.1021/acs.bioconjchem.6b00261 |
| [48] |
E. Nagele, M. Schelhaas, N. Kuder, H. Waldmann, J. Am. Chem. Soc. 120 (1998) 6889-6902. DOI:10.1021/ja9805627 |
| [49] |
L. Brunsveld, J. Kuhlmann, K. Alexandrov, et al., Angew. Chem. Int. Ed. Engl. 45 (2006) 6622-6646. DOI:10.1002/(ISSN)1521-3773 |
| [50] |
E.K. Dolence, J.M. Dolence, C.D. Poulter, Bioconjugate Chem. 12 (2001) 35-43. DOI:10.1021/bc000036g |
| [51] |
G. Kragol, M. Lumbierres, J.M. Palomo, H. Waldmann, Angew. Chem. 116 (2004) 5963-5966. DOI:10.1002/(ISSN)1521-3757 |
| [52] |
J.M. Palomo, M. Lumbierres, H. Waldmann, Angew. Chem. Int. Ed. Engl. 45 (2006) 477-481. DOI:10.1002/(ISSN)1521-3773 |
| [53] |
G. Triola, M. Gerauer, K. Gormer, L. Brunsveld, H. Waldmann, Chemistry 16 (2010) 9585-9591. DOI:10.1002/chem.201001642 |
| [54] |
V. Diaz-Rodriguez, D.G. Mullen, E. Ganusova, J.M. Becker, M.D. Distefano, Org. Lett. 14 (2012) 5648-5651. DOI:10.1021/ol302592v |
| [55] |
A. Yang, Y. Li, S. Pantoom, G. Triola, Y.W. Wu, ChemBioChem 14 (2013) 1296-1300. DOI:10.1002/cbic.v14.11 |
| [56] |
Y.C. Huang, Y.M. Li, Y. Chen, et al., Angew. Chem. Int. Ed. Engl. 52 (2013) 4858-4862. DOI:10.1002/anie.v52.18 |
| [57] |
Y.X. Chen, S. Koch, K. Uhlenbrock, et al., Angew. Chem. Int. Ed. Engl. 49 (2010) 6090-6095. DOI:10.1002/anie.201001884 |
| [58] |
S.Y. Zhang, B. Sperlich, F.Y. Li, et al., ACS Chem. Biol. 12 (2017) 1703-1710. DOI:10.1021/acschembio.7b00165 |
| [59] |
D. Gottlieb, C. Grunwald, C. Nowak, J. Kuhlmann, H. Waldmann, Chem. Commun. (2006), 260-262. |
| [60] |
K. Kuhn, D.J. Owen, B. Bader, et al., J. Am. Chem. Soc. 123 (2001) 1023-1035. DOI:10.1021/ja002723o |
| [61] |
B. Ludolph, H. Waldmann, Chemistry 9 (2003) 3683-3691. DOI:10.1002/chem.200304822 |
| [62] |
H. Mao, S.A. Hart, A. Schink, B.A. Pollok, J. Am. Chem. Soc. 126 (2004) 2670-2671. DOI:10.1021/ja039915e |
| [63] |
A. Dementiev, Protein Exp. Purif. 84 (2012) 86-93. DOI:10.1016/j.pep.2012.04.021 |
| [64] |
D.K. Simanshu, D.V. Nissley, F. McCormick, Cell 170 (2017) 17-33. DOI:10.1016/j.cell.2017.06.009 |
| [65] |
J.R. Silvius, Curr. Top. Membr. 2002 (2002) 371-395. |
| [66] |
B. Bader, K. Kuhn, D.J. Owen, et al., Nature 403 (2000) 223-226. DOI:10.1038/35003249 |
| [67] |
D. Huster, K. Kuhn, D. Kadereit, H. Waldmann, K. Arnold, Angew. Chem. Int. Ed. Engl. 40 (2001) 1056-1058. DOI:10.1002/(ISSN)1521-3773 |
| [68] |
D. Huster, P. Muller, K. Arnold, A. Herrmann, Biophys. J. 80 (2001) 822-831. DOI:10.1016/S0006-3495(01)76061-4 |
| [69] |
D. Huster, A. Vogel, C. Katzka, et al., J. Am. Chem. Soc. 125 (2003) 4070-4079. DOI:10.1021/ja0289245 |
| [70] |
A. Vogel, C.P. Katzka, H. Waldmann, et al., J. Am. Chem. Soc. 127 (2005) 12263-12272. DOI:10.1021/ja051856c |
| [71] |
L. Brunsveld, H. Waldmann, D. Huster, Biochim. Biophys. Acta 1788 (2009) 273-288. DOI:10.1016/j.bbamem.2008.08.006 |
| [72] |
L. Janosi, Z. Li, J.F. Hancock, A.A. Gorfe, Proc. Natl. Acad. Sci. U. S. A. 109 (2012) 8097-8102. DOI:10.1073/pnas.1200773109 |
| [73] |
A. Vogel, G. Reuther, K. Weise, et al., Angew. Chem. Int. Ed. Engl. 48 (2009) 8784-8787. DOI:10.1002/anie.v48:46 |
| [74] |
K. Weise, G. Triola, L. Brunsveld, H. Waldmann, R. Winter, J. Am. Chem. Soc. 131 (2009) 1557-1564. DOI:10.1021/ja808691r |
| [75] |
K. Weise, S. Kapoor, C. Denter, et al., J. Am. Chem. Soc. 133 (2011) 880-887. DOI:10.1021/ja107532q |
| [76] |
J.A. Parker, C. Mattos, Mol. Cancer Res. 13 (2015) 595-603. DOI:10.1158/1541-7786.MCR-14-0535 |
| [77] |
J.T. Swarthout, S. Lobo, L. Farh, et al., J. Biol. Chem. 280 (2005) 31141-31148. DOI:10.1074/jbc.M504113200 |
| [78] |
M. Schmick, A. Kraemer, P.I. Bastiaens, Trends Cell Biol. 25 (2015) 190-197. DOI:10.1016/j.tcb.2015.02.004 |
| [79] |
M. Hanzal-Bayer, L. Renault, P. Roversi, A. Wittinghofer, R.C. Hillig, EMBO J. 21 (2002) 2095-2106. DOI:10.1093/emboj/21.9.2095 |
| [80] |
V. Nancy, I. Callebaut, A. El Marjou, J. de Gunzburg, J. Biol. Chem. 277 (2002) 15076-15084. DOI:10.1074/jbc.M109983200 |
| [81] |
S.A. Ismail, Y.X. Chen, A. Rusinova, et al., Nat. Chem. Biol. 7 (2011) 942-949. DOI:10.1038/nchembio.686 |
| [82] |
A. Chandra, H.E. Grecco, V. Pisupati, et al., Nat. Cell Biol. 14 (2011) 148-158. |
| [83] |
S. Dharmaiah, L. Bindua, T.H. Trana, et al., Proc.Natl. Acad.Sci. U. S. A. 113 (2016) E6766-E6775. DOI:10.1073/pnas.1615316113 |
| [84] |
B. Chen, Y. Jiang, S. Zeng, et al., PLoS Genet. 6 (2010) e1001235. DOI:10.1371/journal.pgen.1001235 |
| [85] |
O. Rocks, M. Gerauer, N. Vartak, et al., Cell 141 (2010) 458-471. DOI:10.1016/j.cell.2010.04.007 |
| [86] |
P.J. Sung, F.D. Tsai, H. Vais, et al., Proc. Natl. Acad. Sci. U. S. A. 110 (2013) 20593-20598. DOI:10.1073/pnas.1306431110 |
| [87] |
T.G. Bivona, S.E. Quatela, B.O. Bodemann, et al., Mol. Cell 21 (2006) 481-493. DOI:10.1016/j.molcel.2006.01.012 |
| [88] |
H. Jang, A. Banerjee, T. Chavan, V. Gaponenko, R. Nussinov, J. Biol. Chem. 292 (2017) 12544-12559. DOI:10.1074/jbc.M117.785063 |
| [89] |
B. Sperlich, S. Kapoor, H. Waldmann, R. Winter, K. Weise, Biophys. J. 111 (2016) 113-122. DOI:10.1016/j.bpj.2016.05.042 |
 2018, Vol. 29
2018, Vol. 29