b Peking-Tsinghua Center for Life Sciences, Peking University, Beijing 100871, China;
c Synthetic and Functional Biomolecules Center, Beijing National Laboratory for Molecular Sciences, Key Laboratory of Bioorganic Chemistry and Molecular Engineering of Ministry of Education, PKU-IDG/McGovern Institute for Brain Research, Peking University, Beijing 100871, China
Membrane voltage is ubiquitous in cell biology. It arises from the selective charge transport across lipid bilayers and regulates diverse physiological processes, with the most famous being the electrical signaling in neurons and cardiomyocytes [1]. While the classical patch-clamp technique has enabled fast and sensitive tracking of membrane voltage at the single cell level, it is difficult to parallelize this technique for recording from a large cell population. In comparison, optical recording methods readily offer high spatial resolution and measurement throughput. For this reason, much effort has been devoted to the development of fluorescent voltage indicators over the past few decades. In particular, genetically-encoded voltage indicators (GEVIs) emerged as promising tools because they allow for cell-specific targeting of measurement. In this mini-review, we outline the development of GEVI designs and highlight their applications in voltage imaging of bioelectric phenomenon. We hope that this article will inform chemical biologists and protein engineers of the history of GEVI development and inspire ideas for future improvements.
A voltage indicator acts as an electrochromic signal transducer. In many cases, the voltage-sensing moiety initially transduces electrical signal into intramolecular mechanical stress, which drives the conformational change of appended fluorescent protein (FP) reporters. This is best exemplified in GEVIs derived from either ion channels or voltage-sensitive phosphatases (VSP). While the detailed mechanism was not well understood, it is generally accepted that voltage sensitivity arises from voltage-induced movement of the fourth transmembrane helix, due to its multiple positively charged amino acid residues. In other cases, the voltagesensing domain could utilize the transmembrane electric field to shift the chemical equilibrium betweenprotonated and deprotonated states of a membrane-anchored retinal chromophore, as is shown in the case of rhodopsin-derived GEVIs. The protonated state is more fluorescent than the deprotonated state, due to stronger absorption in the visible spectrum. Fig. 1 outlines the structure of these designs.

|
Download:
|
| Fig. 1. Design and applications of GEVIs based on voltage-dependent conformational changes. A) The first generation voltage indicator, FlaSh, was built upon voltage-gated potassium channel. B) VSFP2 series are ratiometric reporters consisting of a FRET pair fused to VSD. C) VSFP Butterfly has FPs fused separately to the two termini. D) ArcLight is a monochromic GEVI with sensitive voltage response. E) ASAP1 couples the conformational changes in VSD to cpGFP. F) VSFP2.3 reports membrane voltage transients in hippocampal pyramidal neurons through differential two-color fluorescence imaging. Reprinted with permission [2]. Copyright 2010, Nature Publishing Group. G) Simultaneous recording of multiple neurons with ArcLight in Drosophila brain. Reprinted with permission [3]. Copyright 2013, Elsevier Inc. H) Voltage imaging with ASAP in Drosophila visual system reveals the transformation of voltage responses between pre-synaptic axons and post-synaptic dendrites.Reprinted with permission [4]. Copyright 2016, Elsevier Inc. | |
The first GEVI, called FlaSh, was constructed almost two decades ago as a chimera of voltage-gated Shaker potassium channel and modified green fluorescent protein (GFP [5]).Subsequent mutations in both GFP and the ion channel resulted in spectral variants with voltage sensitivity ranging between 1%-5% ΔF/F per 100 mV (Table 1) [6]. However, the response time constants of these GEVIs typically range from 10-200 ms, which are too slow to capture the millisecond-scale neuronal action potentials. With shorter linker between the FP and the ion channel, response time could reach sub-millisecond range (VSFP1) [7]. Meanwhile, Ataka et al. sought to improve on the kinetics by using voltage-gated sodium channel as the voltage-sensing domain (SPARC) and achieved response time of 2 ms [8]. Coincidentally, these faster sensors have reduced voltage sensitivity, less than 2% ΔF/F per 100 mV. A more recent study based on in silico search identified voltage-gated proton channel from fluke Clonorchissinesis to construct a novel GEVI called Pado [9]. Unfortunately, all of these sensors exhibited modest voltage sensitivity, and many suffered from poor membrane trafficking in mammalian cells [10]. The transmembrane voltage-sensing domain (VSD) of the ascidian Cionaintestinalis voltage-sensing phosphatase (Ci-VSP) represents another important protein scaffold for GEVI design. VSP was initially discovered in 2005 via a genomic survey for ion channel homologues [11]. Following this discovery, a series of GEVIs was constructed by fusing VSD to pairs of FPs that act as Förster resonance energy transfer (FRET) reporters for protein conformational changes. The rationale behind these VSD-FRET sensors was that voltage-dependent conformational changes in the fourth transmembrane helix of the VSD shift the distance and relative orientationbetween the donor and the acceptor, thereby modulating their FRET efficiency. FRET donor and acceptor FPs could be fused to the VSD in two ways: either in tandem at the C-terminus (VSFP2.1 comprising a CFP/YFP pair [12]), or separately at both Nand C-termini (Butterfly [13, 14]). Further improvements include linker optimization [15] and spectral variations (VSFP2.4 [16], Mermaid [17], VSFP-CR [18], Table 1). Compared with ion channelderived GEVIs, there is a remarkable improvement of membrane trafficking in VSD-derived GEVIs, presumably due to the monomeric nature of Ci-VSP.
|
|
Table 1 Summary of GEVIs. |
While many of these FRET sensors have good voltage sensitivity (DR/R approaching 14% per 100 mV) and have been applied to report membrane voltage fluctuations in vivo (optical EEG [13]), they suffer from slow voltage response, particularly during the repolarization step.Response kinetics could be improved by exploring Ci-VSP homologs in other species. For example, replacement of VSD in VSFP2.1 with the VSP genes from sea anemone and zebrafish resulted in GEVIs with 2 ms voltageresponse time constant (Zahra [19]). Another approach is to transplant homologous amino acid motifs from fast voltage-gated potassium channel, Kv3.1, to the VSD [20]. This ion channel-VSD chimera is capable of reporting membrane voltage oscillations up to 200 Hz [21].
While it was initially believed that FRET mechanism was solely responsible for the observed fluorescence changes in VSD-FP pairs, it was discovered later that, at least in some variants, this was not the case. For example, photobleaching of the acceptor YFP in VSFP2A (a variant of VSFP2.1) did not abolish the membrane voltage-dependent fluorescence response in the donor CFP [15]. This observation has inspired the design of a new generation of VSD-based GEVIs with a single FP as the fluorescence reporter (VSFP3.1) [15]. Spectral variants of VSFP3.1 were subsequently generated with different color FP fusions, all of which exhibited similar kinetics and dynamic range [22]. These monochromatic VSD-FP sensors can be more readily paired with other fluorescent sensors for multiplex imaging purposes.
In 2012, Jin et al. reported a VSD-FP mutant with dramatically enhanced voltage sensitivity. Subsequent mutagenesis and linker optimization led to the identification of ArcLight Q239 with voltage sensitivity reaching 35% ΔF/F per 100 mV [23]. The main drawback of ArcLight is its slow temporal response (>10 ms time constant) which low-pass filtered the action potential waveform and reduced the overall spike-detection sensitivity down to 3.2% ΔF/F. To improve the response kinetics, the VSD domain in ArcLightwas replaced with homologues from other species. VSDs from chicken and zebrafish offered faster kinetics [24]. Alternatively, sequence alignment and cassette mutagenesis were employed to improve ArcLight-type sensor. Recently, a triple mutant with truncated linker, called Bongwoori, was identified with this strategy, which resolved 60 Hz action potentials spikes [25].
Circularly permuted FP (cpFP) was introduced as fluorescence reporter of VSD conformational change.This idea was initially tested in 2009 for C-terminal fusion of VSD with cpFP derived from EGFP and far-red mKate, but the resulting GEVIs have slow (>50 ms time constants) and small response ( < 1% ΔF/F per 100 mV) [26]. Subsequent systematic investigations of cpFP hole variations and VSD linker fusion sites identified a fast GEVI, called ElectricPk [27]. A more recent example is the development of a bright red VSDcpmApple GEVI, called FlicR1, from library screen of thousands of mutants. FlicR1 reported action potentials with 3% [11_TD$DIFF]ΔF/F [28]. In addition to C-terminal fusions, cpFPscould also be insertedintothe extracellular loop region. Such design proved to have fast and sensitive response to membrane voltage, as exemplified in ASAP1/ 2f, which detected of spike trains up to 200 Hz [29] and could be applied to imaging action potential in vivo with two-photon illumination [4].
Since 2011, rhodopsins have emerged as a novel protein scaffold for voltage sensing. As a light-sensing protein with seven transmembrane helices and a covalently bound retinal chromophore, rhodopsins often serve as ion pumps or channels that utilize light energy to regulate ion transport across the membrane. In the case of proton-pumping rhodopsins, the Schiff base, which links the retinal to the protein scaffold, could be reversibly protonated and deprotonated during the light-driven ion transport cycle. The protonated state was found to be weaklyfluorescent in the deep red range, and the deprotonated state was not fluorescent. Kralj et al. hypothesized that transmembrane electric field could perturb the local electrochemical potential of protons on the Schiff base, thereby changing the energy landscape of protonation and modulating fluorescence emission. A proteorhodopsin mutant (PROPS) was designed based on this voltage-dependent acid-base equilibrium principleand was successfully applied to probe electrical spiking in bacteria [37]. However, PROPS fails to report membrane voltage in eukaryotic cells, due to lack of plasma membrane localization.
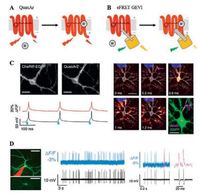
Screening of other microbial rhodopsins identified Archaerhodopsin 3 (Arch) from Halorubrumsodomense as a GEVI that resolves individual action potentials in mammalian neurons in vivo [30]. Arch has sub-millisecond response kinetics and changed fluorescence by ~35% upon 100 mV voltage change. Introducing protonpumping defective mutation (Arch D95N) increases voltage sensitivity by 50%, but slows down voltage response (41 ms). Further mutagenesis near the chromophore binding pocket yields Arch-EEN and Arch-EEQ with improved speed and sensitivity [4]. Directed protein evolution led to the discovery of mutations that improved the brightness, kinetics, and dynamic range of Arch variants (Archers [33, 38] and QuasArs [31]). The red-shifted spectra of Arch-based GEVIs facilitate combination with other GFPbased fluorescent reporters or a variety of optogenetic actuators. For example, fusion of QuasArs and calcium indicator GCaMP6 (CaViar) has enabled simultaneous measurement of calcium and voltage signal in cardiomyocytes [39, 40]. The spectral separation between blue light-activated ion channels and red light-absorbing GEVIs allows for simultaneous optical stimulation and recording. Hochbaum et al. reported an all-optical method, called Optopatch, for studying cellular electrophysiology [31]. By faithfully detecting action potentials, Optopatch measures neuronal excitabilities in cultured neurons, brain slices [31] and mice somatosensory ganglia [41]. The high-throughput nature of optical measurements has enabled screening for drugs affecting ion channels [39, 42]. The Achilles' heel of rhodopsin-derived GEVIs was their dim fluorescence due to low quantum yield. Even with the brightest variants, QuasArs, voltage imaging would require laser illumination at above 200 W/cm2, about two orders of magnitude higher than typical GFP imaging conditions [30, 31] (Fig. 2).
|
Download:
|
| Fig. 2. Design and applications of GEVIs based on voltage-dependent chemical equilibrium between bright and dark states. A) Membrane voltage drives the reversible protonation/deprotonation of retinal chromophore in microbial rhodopsin, changing the native fluorescence. B) Electrochromic FRET is achieved through fusion of a bright fluorescent protein in close proximity to the rhodopsin quencher (also known as FRET-opsin configuration). C) Optopatch enables high-fidelity optical stimulation and recording in rat hippocampal neurons.Action potential propagation within a neuron could be resolved via sub-frame interpolation. Reprinted with permission [31]. Copyright 2014, Nature Publishing Group. D) Voltage imaging of mouse visual cortex with Ace2N-mNeon resolves individual action potentials. Reprinted with permission [36]. Copyright 2015, American Association for the Advancement of Science. | |
To address the brightness issue of Arch-derived GEVIs, one could employ electrochromic FRET (eFRET) mechanism, where voltage-driven acid-base equilibrium led to changes in the rhodopsin absorption spectrum, thereby modulating the degree of energy transfer between an FP donor and the retinal quencher [34]. This FRET-opsin configuration was successfully implemented with two other proton-pumping rhodopsins, derived from fungus Leptosphaeriamaculans and green algae Acetabularia acetabulum, respectively [35, 36]. The broad absorption spectra of these rhodopsins allowed fusion of FPs with different colors, thus leading to a palette of bright and fast GEVIs [34]. Brinks et al. applied eFRET mechanism to measure the absolute membrane voltage scale via two-photon lifetime imaging [43].
To conclude, the development of GEVIs over the past two decades has mainly capitalized on three protein scaffolds with distinct voltage-sensing mechanisms: ion channels and VSPs that undergo voltage-induced conformational changes, and microbial rhodopsins that have voltage-dependent chemical equilibrium. Future improvement would benefit from joint efforts of rational designs based on better understanding of these voltage-sensing mechanisms, as well as high-throughput screening for identifying mutants with better performance.
Acknowledgments
This work was supported by the Peking-Tsinghua Center for Life Sciences and the National Natural Science Foundation of China (No. 21673009). P. Zou was supported by the National Thousand Young Talents Award.
| [1] |
Y. Xu, P. Zou, A.E. Cohen, Curr. Opin. Chem. Biol. 39(2017) 1-10. |
| [2] |
W. Akemann, H. Mutoh, A. Perron, J. Rossier, T. Knöpfel, Nat. Methods 7(2010) 643-649. DOI:10.1038/nmeth.1479 |
| [3] |
G. Cao, J. Platisa, V.A. Pieribone, et al., Cell 154(2013) 904-913. DOI:10.1016/j.cell.2013.07.027 |
| [4] |
H.H. Yang, F.Pierre St-, X. Sun, et al., Cell 166(2016) 245-257. DOI:10.1016/j.cell.2016.05.031 |
| [5] |
M.S. Siegel, E.Y. Isacoff, Neuron 19(1997) 735-741. DOI:10.1016/S0896-6273(00)80955-1 |
| [6] |
G. Guerrero, M.S. Siegel, B. Roska, E. Loots, E.Y. Isacoff, Biophys. J. 83(2002) 3607-3618. DOI:10.1016/S0006-3495(02)75361-7 |
| [7] |
R. Sakai, V. Repunte-Canonigo, C.D. Raj, T. Knopfel, Eur. J. Neurosci. 13(2001) 2314-2318. DOI:10.1046/j.0953-816x.2001.01617.x |
| [8] |
K. Ataka, V.A. Pieribone, Biophys. J. 82(2002) 509-516. DOI:10.1016/S0006-3495(02)75415-5 |
| [9] |
B.E. Kang, B.J. Baker, Sci. Rep. 6(2016) 23865. DOI:10.1038/srep23865 |
| [10] |
B.J. Baker, H. Lee, V.A. Pieribone, et al., J. Neurosci. Methods 161(2007) 32-38. DOI:10.1016/j.jneumeth.2006.10.005 |
| [11] |
Y. Murata, H. Iwasaki, M. Sasaki, K. Inaba, Y. Okamura, Nature 435(2005) 1239-1243. DOI:10.1038/nature03650 |
| [12] |
D. Dimitrov, Y. He, H. Mutoh, et al., PLoS One 2(2007) e440. DOI:10.1371/journal.pone.0000440 |
| [13] |
W. Akemann, H. Mutoh, A. Perron, et al., J. Neurophysiol. 108(2012) 2323-2337. DOI:10.1152/jn.00452.2012 |
| [14] |
H. Tsutsui, Y. Jinno, A. Tomita, et al., J. Physiol. 591(2013) 4427-4437. DOI:10.1113/jphysiol.2013.257048 |
| [15] |
A. Lundby, H. Mutoh, D. Dimitrov, W. Akemann, T. Knopfel, PLoS One 3(2008) e2514. DOI:10.1371/journal.pone.0002514 |
| [16] |
H. Mutoh, A. Perron, D. Dimitrov, et al., PLoS One 4(2009) e4555. DOI:10.1371/journal.pone.0004555 |
| [17] |
H. Tsutsui, S. Karasawa, Y. Okamura, A. Miyawaki, Nat. Methods 5(2008) 683-685. DOI:10.1038/nmeth.1235 |
| [18] |
A.J. Lam, F. St-Pierre, Y. Gong, et al., Nat. Methods 9(2012) 1005-1012. DOI:10.1038/nmeth.2171 |
| [19] |
B.J. Baker, L. Jin, Z. Han, et al., J. Neurosci. Methods 208(2012) 190-196. DOI:10.1016/j.jneumeth.2012.05.016 |
| [20] |
Y. Mishina, H. Mutoh, T. Knopfel, Biophys. J. 103(2012) 669-676. DOI:10.1016/j.bpj.2012.07.031 |
| [21] |
Y. Mishina, H. Mutoh, C. Song, T. Knopfel, Front. Mol. Neurosci. 7(2014) 78. |
| [22] |
A. Perron, H. Mutoh, T. Launey, T. Knopfel, Chem. Biol. 16(2009) 1268-1277. DOI:10.1016/j.chembiol.2009.11.014 |
| [23] |
L. Jin, Z. Han, J. Platisa, et al., Neuron 75(2012) 779-785. DOI:10.1016/j.neuron.2012.06.040 |
| [24] |
Z. Han, L. Jin, J. Platisa, et al., PLoS One 8(2013) e81295. DOI:10.1371/journal.pone.0081295 |
| [25] |
H.H. Piao, D. Rajakumar, B.E. Kang, E.H. Kim, B.J. Baker, J. Neurosci. 35(2015) 372-385. DOI:10.1523/JNEUROSCI.3008-14.2015 |
| [26] |
S.G. Gautam, A. Perron, H. Mutoh, T. Knopfel, Front. Neuroeng. 2(2009) 14. |
| [27] |
L. Barnett, J. Platisa, M. Popovic, V.A. Pieribone, T. Hughes, PLoS One 7(2012) e43454. DOI:10.1371/journal.pone.0043454 |
| [28] |
A.S. Abdelfattah, S.L. Farhi, Y. Zhao, et al., J. Neurosci. 36(2016) 2458-2472. DOI:10.1523/JNEUROSCI.3484-15.2016 |
| [29] |
F. St-Pierre, J.D. Marshall, Y. Yang, et al., Nat. Neurosci 17(2014) 884-889. DOI:10.1038/nn.3709 |
| [30] |
J.M. Kralj, A.D. Douglass, D.R. Hochbaum, D. Maclaurin, A.E. Cohen, Nat. Methods 9(2011) 90-95. DOI:10.1038/nmeth.1782 |
| [31] |
D.R. Hochbaum, Y. Zhao, S.L. Farhi, et al., Nat. Methods 11(2014) 825-833. DOI:10.1038/nmeth.3000 |
| [32] |
Y. Gong, J.Z. Li, M.J. Schnitzer, PLoS One 8(2013) e66959. DOI:10.1371/journal.pone.0066959 |
| [33] |
N.C. Flytzanis, C.N. Bedbrook, H. Chiu, et al., Nat. Commun. 5(2014) 4894. DOI:10.1038/ncomms5894 |
| [34] |
P. Zou, Y. Zhao, A.D. Douglass, et al., Nat. Commun. 5(2014) 4625. |
| [35] |
Y. Gong, M.J. Wagner, J.Li Zhong, M.J. Schnitzer, Nat. Commun. 5(2014) 3674. |
| [36] |
Y. Gong, C. Huang, J.Z. Li, et al., Science 350(2015) 1361-1366. DOI:10.1126/science.aab0810 |
| [37] |
J.M. Kralj, D.R. Hochbaum, A.D. Douglass, A.E. Cohen, Science 333(2011) 345-348. DOI:10.1126/science.1204763 |
| [38] |
R.S. McIsaac, M.K. Engqvist, T. Wannier, et al., Proc. Natl. Acad. Sci. U. S. A. 111(2014) 13034-13039. DOI:10.1073/pnas.1413987111 |
| [39] |
G.T. Dempsey, K.W. Chaudhary, N. Atwater, et al., J. Pharmacol. Toxicol. Methods 81(2016) 240-250. DOI:10.1016/j.vascn.2016.05.003 |
| [40] |
J.H. Hou, J.M. Kralj, A.D. Douglass, F. Engert, A.E. Cohen, Front. Physiol. 5(2014) 344. |
| [41] |
S. Lou, Y. Adam, E.N. Weinstein, et al., J. Neurosci. 36(2016) 11059-11073. DOI:10.1523/JNEUROSCI.1582-16.2016 |
| [42] |
H. Zhang, E. Reichert, A.E. Cohen, Elife 5(2016) e15202. |
| [43] |
D. Brinks, A.J. Klein, A.E. Cohen, Biophys. J. 109(2015) 914-921. DOI:10.1016/j.bpj.2015.07.038 |
 2017, Vol. 28
2017, Vol. 28 



