b Institutes of Nano Electronic Engineering, University Malaysia Perlis, 01000 Kangar, Perlis, Malaysia
Nanotechnology has become increasingly popular because of their unique physical,chemical,optical and catalytic properties compared to their bulk counterparts. Nanotechnology revolutionizes many technologic and industrial sectors and medical instrumentation,homeland security,and many others [1, 2, 3]. Currently,the detection of DNA is an area of great interest as it is the key feature in the research for specific nucleotide sequences. This technique plays an important role in biodiagnostics [4],determination of genetic diversity [5],criminal investigation in forensic and immigration cases [6],food analysis [7, 8] and environmental monitoring [4]. Currently,a variety of high selectivity and sensitivity methods of DNA detection,such as polymerase chain reaction (PCR) [9],terminal-restriction fragment length polymorphis (T-RFLP) [10],chromatography combined with mass spectrometry [11] and surface plasmon resonance (SPR) [12] are widely studied. However,these techniques have their limitations including long assay time,complexity,extensive training and high cost. Therefore,a variety of approaches for the detection of DNA which overcome these weaknesses have been actively investigated,such as electrochemical sensing [13],fluorescence [14],bio-field effect transistor (Bio-FET) [15] and oligonucleotide microarray and DNA sensors [4]. Among these techniques,the electrochemical DNA biosensor with its simplicity,good sensitivity,low cost,and possible miniaturization has attracted significant research interest. Gold nanoparticles (GNPs) have gained considerable attention in recent years for potential applications in nanomedicine due to their interesting size dependent electronics,chemical and optical properties. Also,gold nanoparticles show promise in enhancing the effectiveness of various targeted cancer treatments such as photo thermal therapy and radiotherapy [16]. Nanoparticles have been synthesized by a wide variety of techniques,such as pulsed laser deposition [17],chemical reduction [18],flame metal combustion [19],electrolysis [20],electrochemical reduction [21],solvothermal [22],photo-reduction [23],sono-electrochemical [24],microwave- induced [25],green method [26],aerosol flow reactor [27],spray pyrolysis [28],chemical fluid deposition [29],photochemical reduction [30],and spark discharge [31].
Gold nanoparticles offer some advantages such as high surfacevolume ratio,optical properties,chemical stability and robustness,and has been successfully used in the modification of semiconductor nanostructures for biological diagnostics [32]. GNPs are also widely applied in bio-labeling,self-assembled monolayers (SAM),electron-transfer theories and immunoassays [33, 34] and in various kinds of bimolecular immobilization,such as DNA [7],proteins [35] and enzymes [36].
The detection method relies on the immobilization of single stranded DNA (ssDNA) probes that are complementary and specific for a DNA sequence of the pathogenic target (Fig. 2). GNP probes are used to report the immobilization and hybridization because of their ease of production and functionalization [37, 38]. The nanoparticles conjugated with ssDNA probes specific for the pathogenic target of interest are then hybridized with the DNA test sample,isolated using magnetic separation,and detected through electrochemical analysis [39, 40, 41, 42]. While biosensors using this detection strategy has been shown to detect specific DNA fragments from various pathogens [43, 44, 45],many of these biosensors have only been tested for the detection of purified and gold nanoparticle amplified DNA targets. Similar to some of the limitations of commercially available detection strategies,GNPs are often criticized for its complex,expensive,timeconsuming,and labor-intensive procedural requirements.
Consequently,the need of GNPs for biosensor detection of pathogenic DNA is greatly limited to both field-based and resource limited settings,resulting in the increased need for GNPs in independent biosensor detection methods. The scope of this work,we have developed an easy to fabricate a low cost technique using GNPs/APTES/SiO2/Si/Ti,which acts as the electrode. Immobilizations and hybridizations of DNA were performed using electrochemical detection with potassium hexacyanoferrate to enhance the sensitivity of the DNA detection.
2. Experimental 2.1. Preparation of GNPs solutionGold nanoparticles (GNPs) were used in this project for the immobilization and hybridization of the DNA on the SiO2 thin films. First,a HAuCl4 solution at a concentration of 0.49 mol/L was prepared by dissolving 500 mg of HAuCl4 in 3 mL of 10% HCl. Then,a dilute 0.2 mmol/L of HAuCl4 solution was made by adding 40 mL (19.6 mmol) of HAuCl4 solution to 100 mL of deionized water to produce solution A. Second,558.79 mg of trisodium citrate was added to 50 mL of deionized water to make solution B. The concentration of the solution was controlled at 38.8 mmol/L. Solution A was brought to a rolling boil at 150 ℃ with vigorous stirring to yield a solution of homogenous sized GNPs. Then 10 mL of 38.8 mmol/L of sodium citrate was added rapidly into the vortex of the solution. The added solution resulted in a color change from pale yellow to red. Boiling and stirring was continued for another 10 min. The heating was then removed,and stirring was continued for an additional 15 min. When the solution cooled to room temperature (r.t.),it was filtered through a 0.8 mmmembrane filter paper. The prepared solution was kept under refrigeration at a temperature of 4 ℃ and analyzed by using UV-vis with the wavelength 400 nm to 800 nm,TEM and AFM.
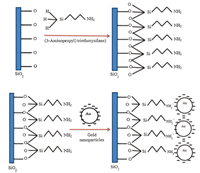
2.2. Modification of SiO2 with GNPsA p-type silicon (1 0 0) wafer (1 cm × 1 cm) was ultrasonically cleaned by using acetone and isopropanol for about 15 min,and then was immersed in the buffered oxide etching (BOE) solution for 30 min and washed with deionized water following the oxidation process. After oxidation,the silicon oxide (SiO2) layer of thickness was ~50 nm,and titanium was deposited on the backside of the Si using thermal evaporator. The selectivity of the DNA biosensor was studied using the GNPs/APTES/SiO2/Si/Ti electrode. The SiO2 surface was functionalized with an APTES solution which was prepared by the mixing of 2% APTES with 93% of ethanol and 5% of deionized water. The silanyl group (-SiH3) presented in APTES was used for the process of silanization,which was chemically attached to the hydroxyl-rich SiO2 [46]. Additionally,the amino group (NH2-) presented in APTES served as a glue layer to attach the GNPs which linked to probe DNA. Attachment between APTES and GNPs is shown in Fig. 1. For the surface modification of SiO2 with APTES,10 μL of prepared APTES solution was placed on the SiO2 surface and incubated for 2 h. The surface was washed 3 times,and blow dried and then 10 mL of GNPs was dropped on the surface at 150 ℃ for 20 min on hot plate. This step was repeated 3 times to obtain enough GNPs on the SiO2 surface and to prepare the electrode for electrical characterization.
|
Download:
|
| Fig. 1. Surface modification of SiO2 with GNPs using APTES. | |
2.3. Probe DNA immobilization on modified GNPs
The oligonucleotides used in this project were purchased from 1st BASE Pte Ltd. (Malaysia) and are shown in Table 1. Probe DNA,defined in Table 1,was dropped onto the GNP modified SiO2 electrode for immobilization and incubated at r.t. for 2 h. After 10 min,the electrode was carefully washed using deionized water to remove any un-bonded DNA probe material and dried at r.t. The probe modified devices were denoted as DNA/GNPs/APTES/SiO2/ Si/Ti,and were then ready for electrochemical measurements.
|
|
Table 1 Oligonucleotides used for immobilization and hybridization |
2.4. Target DNA hybridization detection
Hybridization of DNA used in this project was purchased from 1st BASE Pte Ltd. (Malaysia) and are shown in Table 1. To hybridize DNA,10 μL of 10 μmol/L complementary DNA is dropped onto the GNP electrode and incubated for 2 h. After that,the GNP electrode is washed using deionized water to remove any non-hybridized DNA,and dried at r.t. A 10 mL aliquot of 0.5 mmol/L methylene blue is then dropped onto the GNP electrode and incubated again for 3 min. Finally,the GNP electrode is washed once again with deionized water to remove any excess of methylene blue and the GNP electrode is ready to be electrical measured once it has dried.
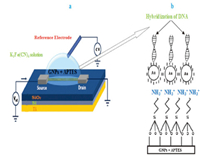
2.5. Electrochemical characterization by cyclic voltammetryElectrochemical measurements were performed by using a dielectric analyzer. The tests were conducted by using Ag/AgCl as the reference electrode and a GNP-modified electrode as a working electrode. The Ti layer acts as a back gate. The responses of the DNA immobilization and hybridization were investigated in 10 mmol/L potassium hexacyanoferrate Ⅲ,K3Fe(CN)6 aqueous solution containing 0.1 mol/L KCl as electrolyte. A schematic view of testing measurement for DNA detection is shown in Fig. 2.
|
Download:
|
| Fig. 2. A schematic illustration of testing measurement (a) a modified GNPs electrode (b) The hybridization DNA on the GNPs using APTES. | |
2.6. Characterization
The morphology of the GNPs was characterized using a transmission electron microscopy (TEM,Libra 120-Carl Zeiss). Ultraviolet-visible (UV-vis-NIR,Perkin Elmer lambda 950) spectroscopy was then used to study the optical properties of the GNPs at r.t. and atomic force microscopy (AFM) was used to study the structural properties of the GNPs. The DNA immobilization and hybridization was tested using a dielectric analyzer (Alpha-A High Performance Frequency Analyzer,Novocontrol Technologies,and Germany).
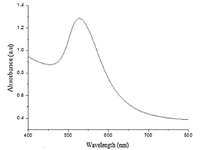
3. Results and discussion 3.1. Measurement of UV-vis spectroscopyThe characterization of prepared solution of GNPs was examined using UV-vis spectroscopy. The measurements were taken over the wavelength range of 400-800 nm under ambient conditions and the result shown in Fig. 3. The absorbance maximum was found at 530 nm,which was indicative of GNPs of diameter 30 ± 5 nm [47]. The particle-size was further confirmed using the method described by Haiss et al. [48] and is shown in the following equation:
|
Download:
|
| Fig. 3. UV–vis absorption spectrum of Au-nanoparticles solution | |
| $d = \frac{{\ln \left( {{\lambda _{{\text{spr}}}} - {\lambda _0}} \right)/{L_1}}}{{{L_2}}}$ | (1) |
where: d = particle diameter; λspr = peak position of Au-nanoparticles,λo = 512,L1 = 6.53,L2 = 0.0216. From Eq. (1),the obtained nanoparticles size was around 28 nm in diameter.
3.2. Characterization of surface morphologyMorphology and microstructure of the GNPs were investigated by transmission electron microscopy (TEM). Fig. 4 shows typical surface morphologies of GNPs as obtained from TEM. The GNPs can be clearly recognized without showing any preferred direction. The GNPs have a structure which is a typical feature of relatively 20-30 nm in diameter.

|
Download:
|
| Fig. 4. TEM Image 100 nm (A) and 200 nm (B) of prepared GNPs | |
3.3. Atomic force microscopy (AFM)
AFM demonstrated the particle size and structure of GNPs. Fig. 5 shows the AFM images which present a two-dimensional (2-D) and three-dimensional (3-D) view of the surface structure of the GNPs. The images confirmed that the GNPs have a surface roughness of 228.89 nm and a small particles size distribution.

|
Download:
|
| Fig. 5. AFM images of 2-D (A) and 3-D (B) of GNPs. | |
3.4. Capacitance measurement
Previous studies documented that the dielectric properties of ssDNA is different from those of dsDNA,especially in the low frequency region [49]. Therefore,we proceeded to detect target DNA hybridization through the measurement of dielectric properties of the Au-modified SiO2 thin films. The change in capacitance before and after hybridization of the DNA at different frequencies was measured using a dielectric analyzer. The measurements were carried out at a frequency range of 1 Hz to 1 MHz. Before the detection of DNA targets,dielectric properties of Au-doped SiO2 thin films were measured to draw the base line for the further detection of DNA targets. The capacitance measurement was also performed when the ssDNA-probe was immobilized onto the Au-doped SiO2 thin films. For hybridization,complementary target DNA was tested on the same device. The results are demonstrated in Fig. 6. It was clearly observed that the capacitance values of the bare,GNP-modified surface and immobilization and target DNA hybridization were 47 × 10-12 F,41 × 10-8 F,21 μF and 14 μF,respectively,at 1 Hz. The capacitance value for the GNPmodified surface is higher than the bare device. The immobilization and hybridization of DNA was successfully detected by showing the highest capacitance values of 21 μF and 14 μF,respectively,on the modified GNP electrodes. The capacitance value between GNP-modified surface and DNA immobilization and hybridization presented the successful use GNPs on thermally oxidized SiO2 thin film for DNA detection. Vetrone et al. [50] investigated the ability of a gold nanoparticle-DNA (AuNP-DNA) biosensor to detect non-PCR amplified genomic Salmonella enterica serovar Enteritidis (S. enteritidis) DNA,from pure or mixed bacterial culture and spiked liquid matrices. Non-PCR amplified DNA was hybridized into sandwich-like structures (magnetic nanoparticles/ DNA/AuNPs) and analyzed through detection of gold voltammetric peaks using differential pulse voltammetry. Reyes et al. [51] reported on a ZnO-nanostructure-based quartz crystal microbalance (nano-QCM) device for biosensing applications. The ZnO nano tips are directly grown on the sensing area of a conventional QCM by metalorganic chemical vapor deposition (MOCVD). The selective immobilization and hybridization of DNA oligonucleotide molecules are confirmed by fluorescence microscopy of the nano- QCM sensing areas.

|
Download:
|
| Fig. 6. Capacitance versus frequency curves of Au-modified SiO2 thin films for DNA immobilization and hybridization detection. | |
3.5. Permittivity measurement
The permittivity measurements were also performed on the same device as shown in Fig. 7. These measurements have the same direction with the capacitance measurement,whereby it gives the largest changes in permittivity with complementary target and probe DNA immobilization. However,it clearly demonstrated that permittivity measurement starts to increase dramatically from 343 × 101 to 193 × 103 and 155 × 103 at the frequency range of ~200 Hz to 1 Hz for bare and DNA immobilization and hybridization,respectively,whereas the capacitance measurements profile started to significantly increase from a frequency of ~1 Hz and tend as the frequency increases. The result revealed that permittivity measurement provides more sensitivity at lower frequency during hybridization. This work demonstrated the changes in capacitance and permittivity values of the GNPmodified electrode during probe DNA immobilization and hybridization confirming the presence of the DNA during the measurement using GNPs electrode.

|
Download:
|
| Fig. 7. Permittivity versus frequency curves of Au-modified SiO2 thin films for DNA immobilization and hybridization detection. | |
3.6. Conductivity measurement
Conductivity measurements were also carried out to investigate the effect of probe DNA immobilization and target DNA hybridization on the GNP-modified SiO2 thin films. The measured conductivity values for bare,the GNP-modified SiO2,immobilized and hybridization device were (3.9 × 10-13,2.1 × 10-9,1.2 × 10-7 and 1.1 × 10-7) S-cm-1,respectively. It can be observed from Fig. 8 that probe DNA immobilization and target DNA hybridization on the GNP-modified SiO2 thin films,conductivity was increased on the bare electrode; therefore the resistivity of the device was decreased. This might be due to a strong interaction which occurred between potassium hexacyanoferrate Ⅲ,K3Fe(CN)6 and the unpaired guanine base in the probe DNA [52]. Furthermore,the differences in value of conductivities between GNPs-modified surface and probe DNA immobilization and target DNA hybridization demonstrated the electron transfer occurred during the immobilization of DNA and target DNA hybridization. Therefore,the conductivity measurement confirmed the reaction of the DNA immobilization and target DNA hybridization onto GNPs-modified surface were realized in the electrolyte solution.

|
Download:
|
| Fig. 8. Conductivity versus frequency curves of Au-modified SiO2 thin films for DNA immobilization and hybridization detection. | |
4. Conclusion
A DNA biosensor was successfully fabricated using goldnanoparticle modified SiO2 thin films with IDE electrodes. The gold-nanoparticles were synthesized through a low-cost and easily manipulated technique and GNP modification on SiO2 thin films was performed using a low-cost doping process. The fabricated biosensor successfully differentiated the detection of DNA immobilization and hybridization through the measurement of dielectric and conductivity properties in a blinded,label free approach,suggesting its potential utility in low-cost biodiagnostics,forensic testing,food analysis and environmental monitoring.
| [1] | D. Imre. Titanium dioxide and gold nanoparticle for environmental and biological application. Ann. Fac. Eng. Hunedoara 1 (2011) 161–166 |
| [2] | S.R. Boddu, V.R. Gutti, T.K. Ghosh, R.V. Tompson, S.K. Loyalka. Gold, silver, and palladium nanoparticle/nano-agglomerate generation, collection, and characterization. J. Nanopart. Res. 13 (2011) 6591–6601 |
| [3] | S.B. Gayathri, P. Kamaraj. Development of electrochemical DNA biosensors-a review. Chem. Sci. Trans. 4 (2015) 303–311 |
| [4] | M.E. Ali, M. Kashif, K. Uddin, et al. Species authentication methods in foods and feeds:the present, past, and future of halal forensics. Food Anal. Methods 5 (2012) 935–955 |
| [5] | L. Hood, D. Galas. The digital code of DNA. Nature 421 (2003) 444–448 |
| [6] | M.J. Heller. DNA microarray technology:devices, systems, and applications. Annu. Rev. Biomed. Eng. 4 (2002) 129–153 |
| [7] | A. Zinchenko, Y. Taki, V.G. Sergeyev, S. Murata. DNA-assisted solubilization of carbon nanotubes and construction of DNA-MWCNT cross-linked hybrid hydrogels. Nanomaterials 5 (2015) 270–283 |
| [8] | M.E. Ali, U. Hashim, S. Mustafa, Y.B. Che Man, T. Adam, Q. Humayun. Nanobiosensor for the detection and quantification of pork adulteration in meatball formulation. J. Exp. Nanosci. 9 (2014) 152–160 |
| [9] | K. Yoshioka, S. Kakumu, T. Wakita, et al. Detection of hepatitis C virus by polymerase chain reaction and response to interferon-α therapy:relationship to genotypes of hepatitis C virus. Hepatology 16 (1992) 293–299 |
| [10] | A.M. Osborn, E.R.B. Moore, K.N. Timmis. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2 (2000) 39–50 |
| [11] | P.B. Farmer, K. Brown, E. Tompkins, et al. DNA adducts:mass spectrometry methods and future prospects. Toxicol. Appl. Pharmacol. 207 (2005) 293–301 |
| [12] | K.M. Byun, N.H. Kim, Y.H. Ko, J.S. Yu. Enhanced surface plasmon resonance detection of DNA hybridization based on ZnO nanorod arrays. Sens. Actuators, B:Chem. 155 (2011) 375–379 |
| [13] | W. Zhang, T. Yang, D.M. Huang, K. Jiao. Electrochemical sensing of DNA immobilization and hybridization based on carbon nanotubes/nano Zinc oxide/chitosan composite film. Chin. Chem. Lett. 19 (2008) 589–591 |
| [14] | D.W. Selinger, K.J. Cheung, R. Mei, et al. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18 (2000) 1262–1268 |
| [15] | C.Y. Hsiao, C.H. Lin, C.H. Hung, et al. Novel poly-silicon nanowire field effect transistor for biosensing application. Biosens. Bioelectron. 24 (2009) 1223–1229 |
| [16] | V. Kattumuri, Gold Nanoparticles for Biomedical Applications:Synthesis, Characterization, In Vitro and In Vivo Studies, University of Missouri-Columbia, Columbia, MO, 2006. |
| [17] | T. Donnelly, S. Krishnamurthy, K. Carney, N. McEvoy, J. Lunney. Pulsed laser deposition of nanoparticle films of Au. Appl. Surf. Sci. 254 (2007) 1303–1306 |
| [18] | C.L. Wu, X.L. Qiao, J.G. Chen, et al. A novel chemical route to prepare ZnO nanoparticles. Mater. Lett. 60 (2006) 1828–1832 |
| [19] | S.S. Yang, Y.H. Jang, C.H. Kim, et al. A flame metal combustion method for production of nanoparticles. Powder Technol. 197 (2010) 170–176 |
| [20] | M. Szymańska-Chargot, A. Gruszecka, A. Smolira, J. Cytawa, L. Michalak. Massspectrometric investigations of the synthesis of silver nanoparticles via electrolysis. Vacuum 82 (2008) 1088–1093 |
| [21] | P.Y. Lim, R.S. Liu, P.L. She, C.F. Hung, H.C. Shih. Synthesis of Ag nanospheres particles in ethylene glycol by electrochemical-assisted polyol process. Chem. Phys. Lett. 420 (2006) 304–308 |
| [22] | M.J. Rosemary, T. Pradeep. Solvothermal synthesis of silver nanoparticles from thiolates. J. Colloid Interface Sci. 268 (2003) 81–84 |
| [23] | H.Y. Jia, J.B. Zeng, W. Song, J. An, B. Zhao. Preparation of silver nanoparticles by photo-reduction for surface-enhanced Raman scattering. Thin Solid Films 496 (2006) 281–287 |
| [24] | Y.C. Liu, L.H. Lin, W.H. Chiu. Size-controlled synthesis of gold nanoparticles from bulk gold substrates by sonoelectrochemical methods. J. Phys. Chem. B 108 (2004) 19237–19240 |
| [25] | J.L. Gu, W. Fan, A. Shimojima, T. Okubo. Microwave-induced synthesis of highly dispersed gold nanoparticles within the pore channels of mesoporous silica. J. Solid State Chem. 181 (2008) 957–963 |
| [26] | H.Z. Huang, X.R. Yang. Synthesis of polysaccharide-stabilized gold and silver nanoparticles:a green method. Carbohydr. Res. 339 (2004) 2627–2631 |
| [27] | H. Eerikäinen, E.I. Kauppinen. Preparation of polymeric nanoparticles containing corticosteroid by A novel aerosol flow reactor method. Int. J. Pharm. 263 (2003) 69–83 |
| [28] | Y. Itoh, M. Abdullah, K. Okuyama. Direct preparation of nonagglomerated indium tin oxide nanoparticles using various spray pyrolysis methods. J. Mater. Res. 19 (2004) 1077–1086 |
| [29] | M. Duocastella, J.M. Fernández-Pradas, J. Domínguez, P. Serra, J.L. Morenza. Printing biological solutions through laser-induced forward transfer. Appl. Phys. A 93 (2008) 941–945 |
| [30] | K.L. McGilvray, M.R. Decan, D.S. Wang, J.C. Scaiano. Facile photochemical synthesis of unprotected aqueous gold nanoparticles. J. Am. Chem. Soc. 128 (2006) 15980–15981 |
| [31] | N.S. Tabrizi, M. Ullmann, V.A. Vons, U. Lafont, A. Schmidt-Ott. Generation of nanoparticles by spark discharge. J. Nanopart. Res. 11 (2009) 315–332 |
| [32] | A. Matsumoto, N. Sato, K. Kataoka, Y. Miyahara. Noninvasive sialic acid detection at cell membrane by using phenyl boronic acid modified self-assembled monolayer gold electrode. J. Am. Chem. Soc. 131 (2009) 12022–12023 |
| [33] | M.M. Rahman, X.B. Li, N.S. Lopa, S.J. Ahn, J.-J. Lee. Electrochemical DNA hybridization sensors based on conducting polymers. Sensors 15 (2015) 3801–3829 |
| [34] | J.L. Qu, L.D. Wu, H. Liu, X.C. Fu, Y. Song. A novel electrochemical biosensor based on DNA for rapid and selective detection of cadmium. Int. J. Electrochem. Sci. 10 (2015) 4020–4028 |
| [35] | J.M. Abad, S.F.L. Mertens, M. Pita, V.M. Fernández, D.J. Schiffrin. Functionalization of thioctic acid-capped gold nanoparticles for specific immobilization of histidine-tagged proteins. J. Am. Chem. Soc. 127 (2005) 5689–5694 |
| [36] | S. Phadtare, V.P. Vinod, K. Mukhopadhyay, A. Kumar, M. Rao. Immobilization and biocatalytic activity of fungal protease on gold nanoparticle-loaded Zeolite microspheres. Biotechnol. Bioeng. 85 (2004) 629–637 |
| [37] | K. Saha, S.S. Agasti, C. Kim, X.N. Li, V.M. Rotello. Gold nanoparticles in chemical and biological sensing. Chem. Rev. 112 (2012) 2739–2779 |
| [38] | G. Doria, J. Conde, B. Veigas, L. Giestas, C. Almeida, et al. Noble metal nanoparticles for biosensing applications. Sensors 12 (2012) 1657–1687 |
| [39] | D. Zhang, M.C. Huarng, E.C. Alocilja. A multiplex nanoparticle-based bio-barcode DNA sensor for the simultaneous detection of multiple pathogens. Biosens. Bioelectron. 26 (2010) 1736–1742 |
| [40] | T.G. Drummond, M.G. Hill, J.K. Barton. Electrochemical DNA sensors. Nat. Biotechnol. 21 (2003) 1192–1199 |
| [41] | J. Wang. Portable electrochemical systems. TrAC Trends Anal. Chem. 21 (2002) 226–232 |
| [42] | J. Weng, J.F. Zhang, H. Li, et al. Label-free DNA sensor by boron-doped diamond electrode using an AC impedimetric approach. Anal. Chem. 80 (2008) 7075–7083 |
| [43] | E.D. Goluch, J.M. Nam, D.G. Georganopoulou, et al. A bio-barcode assay for onchip attomolar-sensitivity protein detection. Lab Chip 6 (2006) 1293–1299 |
| [44] | J.M. Nam, C.S. Thaxton, C.A. Mirkin. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science 301 (2003) 1884–1886 |
| [45] | J.M. Nam, S.I. Stoeva, C.A. Mirkin. Bio-bar-code-based DNA detection with PCRlike sensitivity. J. Am. Chem. Soc. 126 (2004) 5932–5933 |
| [46] | X.M. Hou, L.X. Wang, G.F. He, J.C. Hao. Synthesis, optical and electrochemical properties of ZnO nanorod hybrids loaded with high-density gold nanoparticles. CrystEngComm 14 (2012) 5158–5162 |
| [47] | M.E. Ali, U. Hashim, S. Mustafa, et al. Nanoparticle sensor for label free detection of swine DNA in mixed biological samples. Nanotechnology 22 (2011) 195503–195510 |
| [48] | W. Haiss, N.T.K. Thanh, J. Aveyard, D.G. Fernig. Determination of size and concentration of gold nanoparticles from UV-vis spectra. Anal. Chem. 79 (2007) 4215–4221 |
| [49] | P.E. Canavar, E. Ekşin, A. Erdem. Electrochemical monitoring of the interaction between mitomycin C and DNA at chitosan-carbon nanotube composite modified electrodes. Turk. J. Chem. 39 (2015) 1–12 |
| [50] | S.A. Vetrone, M.C. Huarng, E.C. Alocilja. Detection of non-PCR amplified S. enteritidis genomic DNA from food matrices using a gold-nanoparticle DNA biosensor:a proof-of-concept study. Sensors 12 (2012) 10487–10499 |
| [51] | P.I. Reyes, Z. Zhang, H.H. Chen, et al. A ZnO nanostructure-based quartz crystal microbalance device for biochemical sensing. IEEE Sens. J. 9 (2009) 1302–1307 |
| [52] | M. Das, G. Sumana, R. Nagarajan, B.D. Malhotra. Zirconia based nucleic acid sensor for Mycobacterium tuberculosis detection. Appl. Phys. Lett. 96 (2010) 133703–133712 |
 2016, Vol. 27
2016, Vol. 27 



