b University of Chinese Academy of Sciences, Beijing 100049, China
In recent years,G-quadruplexes (G4s) have attracted intensive interest due to their biological functions,such as gene expression, gene regulation,and potential as antitumor therapeutic targets [1, 2, 3]. G4s are defined by layers of stacked G-tetrads,each of which include four guanine bases associated through Hoogsteen bonding interactions in the presence of monovalent cations,such as K+ and Na+ [4]. Bioinformatics analysis suggests that G-rich sequences capable of forming G4s are widespread in key regulatory regions of the human genome,such as telomeres [5] and promoters [6]. Functional G4 ligands could recognize and stabilize G-quadruplexes,which plays an important role in designing cancer drugs and diagnosis of various diseases. Specific recognition of G4s from other DNA motifs is an important task in developing necessary G4 ligands.
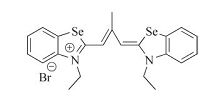
The fundamental unit of G4s is the guanine tetrad,and the large planar surface of terminal G4 can be considered as a characteristic hallmark common to all G4s structures [7]. So far,most reported G4 ligands,such as cyanine dyes [8, 9, 10, 11],cationic macrocycles [12],metal complexes [13, 14],and quinazoline [15] and anthraquinone derivatives [16] are based on heteroaromatic systems π-π stacking binding. However,solely recognizing this shared structural feature is unlikely to confer selectivity due to the diversity of G4s structures. It has been reported that a higher selectivity for G4 s ligands may be achieved by recognizing different groove and loop features among different G4s topologies [17]. Thus,it could be rationally inferred that ligands,which could bind to G4 on the terminal G-quartet and corresponding groove at the same time, might result in high specificity probes. Unfortunately,such ligands are rarely reported [18]. In our previous work,we reported that cyanine dye DMSB (Fig. 1),2,2′-diethyl-9-methyl-selenacarbocyanine bromide,interacted with intermolecular parallel-stranded G4 [d (TGGGGT)]4 (abbreviated TG4T) by dual-site simultaneous binding mode,occupying the 5′-end terminal G-tetrad and the corresponding groove [19].
|
Download:
|
| Fig. 1.The chemical structure of DMSB. | |
In this paper,the recognition ability of DMSB to various DNA motifs will be discussed. This probe also can simply distinguish the parallel-stranded G4s from other DNA motifs by using the UV-vis spectrum.
2. Experimental
The cyanine dye DMSB was synthesized according to Hamer’s [20] and Brooker’s [21] methods. The structure was verified by mass spectrometry and nuclear magnetic resonance (NMR) (S (a) part). All oligonucleotides were purchased from Sangon Biotech Co.,Ltd. (Shanghai,China) and purified by HPLC (purity 98%). Calf thymus DNA (CT) was purchased from Sigma Inc. (D4522). Analytical-grade methanol,KH2PO4,K2HPO4,and EDTA were purchased from Beijing Chem. Co.,(China). Ultrapure water prepared by Milli-Q Gradient ultrapure water system (Millipore) was used throughout the experiments.
The stock solutions of 100 μmol/LDMSB (Fig. 1)were prepared by dissolving them in methanol and then storing in the dark at -4℃ to prevent photodegradation. All oligonucleotides are listed in the S (b) part of SI. The stock solutions of TG4T (Oxytricha telomere),c-myc-2345 (Oncogenic promoter),c-kit1 (Oncogenic promoter),H7 (Human telomere),TBA (Thrombin aptamer),H22 (Human telomere),M24 (sequence human telomeres,which is considered as a mixture of several different quadruplex forms, whose exact structure is still in debate),S17 (non-telomeric, single-stranded DNA),D26 (intramolecular hairpin duplex, synthesis),and CT were prepared by dissolving them directly in PBS (20 mmol/L KH2PO4/K2HPO4,1 mmol/L EDTA,pH 7.4), filtering through a microfiltration membrane (φ = 0.22 μmol/L), and heating to 90℃ for 5 min followed by gradually cooling to room temperature at a rate of 1 ℃min-1. The concentrations of DNA stock solutions were determined based on their absorbance at 260 nm. The stock solutions of double-stranded ds (H22),ds (M24),and ds (AT10) were prepared by heat denaturing and annealing the mixture of the two complementary oligonucleotides at 1:1 molar ratio. All DNA samples were stored for more than 24 h at 4℃ and then structurally identified by circular dichroism (CD) spectra.
Absorption spectra were measured by an Agilent-8453 UV/ visible spectrophotometer in 10 mm quartz cells at room temperature equipped with a Peltier effect heated cuvette holder.
CD spectra (220-400 nm) were recorded on a Jasco J-815 circular dichroism spectrophotometer in 10 mm path-length quartz cells at room temperature. All spectra were collected at a scan speed of 500 nm/min and response time of 0.5 s with five scans averaged.
3. Results and discussion3.1. The binding of DMSB with TG4T
Due to the extended planarπ-electron conjugated system, cyanine dye DMSB (Fig. 1) exists in the form of either monomer or various self-assemblies in different environments. Fig. 2a shows that cyanine dye DMSB presents a primary absorption band at 553 nm assigned to monomer [22] (termed as M-band) in PBS (20 mmol/L K2HPO4/KH2PO4,1 mmol/L EDTA,pH 7.4),as well as a small shoulder absorption band around 512 nm assigned to dimer [23, 24] (termed as D-band) according to the McRaeKasha exciton model [25] and two reported DMSB analogs,DTC [26] and Cy3 [27].

|
Download:
|
| Fig. 2.(a) The absorption spectra of 10μmol/L DMSB in methanol and 20 mmol/L PBS, respectively. (b) Absorption spectra of 12 μmol/L DMSB in the presence of different concentrations of TG4T in 20 mmol/L PBS. | |
[d(TGGGGT)]4 (termed as TG4T),which is a telomeric DNA sequence,could spontaneously form an intermolecular parallel G4 [28] in PBS. With different equivalents of TG4T (from 0.5 mmol/L to 24 μmol/L) to 12 mmol/L DMSB (Fig. 2b),the interaction between DMSB and TG4T presents two stages during the process of titration: dimer rising/monomer falling stage and dimer falling/ monomer rising stage. This result strongly suggests there are two different binding modes in the interaction between TG4T and DMSB,called D-mode (stage of monomer assembling to dimer) in the case of [DMSB]/[TG4T]>3 and M-mode (stage of dimer disassembling to monomer) in the case of [DMSB]/[TG4T] < 3. Our group has revealed that DMSB adopt a dual-site simultaneous binding mode of TG4T [19]. These results suggest that in M-mode DMSB stacks on the 30-end of TG4T in the form of monomer, whereas in D-mode,DMSB could bind to the G-quadruplex in two different modes simultaneously in the form of monomer and groove-binding in dimer. The separation ratio point of the two modes in Fig. 2b is [DMSB]/[TG4T] = 3,which means one TG4T could combine up to three DMSB molecules. This finding also supports the existence of the two binding modes at the same time in D-mode.
3.2. The selectivity of DMSB versus different DNA motifs
Based on the dual binding characteristics of DMSB to TG4T, DMSB could assemble into a cofacial dimer templated by the groove of G4 . Considering that the steric feature of the G4 groove is different from that of the minor groove in duplex DNA [29, 30], DMSB dimer might only form in the presence of specific G4 motifs. Thus,the existence of DMSB dimer or the distribution of DMSB between monomer and dimer could be considered as a signature to verify specific G4s.
To intuitively describe the distribution of DMSB between monomer and dimer,the ratio of absorption maximum of the DMSB M-band to that of the D-band in the presence of certain DNA sample as a distribution coefficient (dc) is defined,shown in Eq. (1).
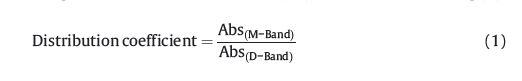
Fig. 3a illustrates the changes of the value of the distribution coefficient for TG4T with the ratio of [DMSB]/[TG4T]. In D-mode, the value of the distribution coefficient of DMSB for TG4T decreases with the decrease of the ratio,whereas in M-mode,it rises remarkably. Thus,the distribution coefficient could be used quantitatively to describe the dimer inducement ability of certain DNA samples. A lower distribution coefficient value represents stronger dimer inducement ability. Fig. 3a shows a dimer domination stage (distribution coefficient is less than 1.0) in the 10-3 range of [DMSB]/[DNAs] molar ratio.

|
Download:
|
| Fig. 3.(a) The distribution coefficient values for TG4T as a function of [DMSB]/[DNAs] molar ratio. (b) Distribution coefficient values for the 13 DNA samples as a function of [DMSB]/[DNAs] molar ratio or mass concentration for CT. | |
To judge the verification ability of DMSB against specific G4 motifs,the DMSB distribution coefficients for another 12 DNA samples with various secondary structures [SI,S (b),S (c) and S (d) part] were measured. As shown in Fig. 3b,in dimer domination stage (molar ratio of [DMSB]/[DNA] in the range of 10-3),the DMSB distribution coefficients for DNA could be grouped into two categories. For parallel G4 s,including intramolecular (c-myc-2345 and c-kit1) and intermolecular ones (H7 and TG4T),the distribution coefficients are relatively low (<1.2). This finding indicates that parallel G4s could provide proper groove cavities; templates on which DMSB could assemble to cofacial dimers. On the other hand,for DNAs with other motifs,including linear duplex CT,ds (H22),and ds (AT10),hairpin duplex D26,single-strand S17, antiparallel G4 (TBA),hybrid G4 (H22),and mixed G4 (M24),the distribution coefficients are higher than those of parallel G4s (almost >1.4). This result indicates that these DNA motifs do not have the proper groove conformation and cannot induce DMSB to assemble into dimers. In addition,our previous data have shown that DMSB could stabilize parallel c-myc-2345 G4 while antiparallel- stranded H22 G4 and duplex CT nearly could not,indicating that dual-site binding mode of DMSB also resulted in higher affinity to parallel G4 [19].
DMSB can recognize parallel G4s (regardless of intermolecular or intramolecular influences and no matter whether the sequences are derived from human telomeres or oncogenic promoters) from other DNA motifs by the formation of dimers in a proper molar ratio of [DMSB]/[DNA] (10 > [DMSB]/[DNA]> 3). The value of the distribution coefficient can be considered as a unique recognizing signature.
Further,we compare recognition ability of specific G4s by the dual-site binding mode of end-stacking. The known probe through end-stacking binding mode [31] is fusion of a thiazole orange and isaindigotone skeleton together. The value of A600/A535 was used to reflect the recognizing ability of distinct DNA motifs,with the absorbance at 535 nm (A535) assigned to H-aggregates and 600 nm (A600) assigned to monomer. The different distributions for the value of A600/A535 could distinguish the G4s (>1.4) from the nonquadruplexes (<0.9). Other probes utilizing end-stacking,such as ETC [9],designed by Yang et al. could recognize the hybrid/ intramolecular G4s from DNA with other motifs by the absorption changes of monomer. In the case of DMSB,the distribution of value monomer to dimer could distinguish parallel G4s from other DNA motifs. The above results illustrated that dual-site binding mode of DMSB exhibited higher selectivity to specific G4s from other DNA conformations than those of ligands by the end-stacking mode.
4. Conclusion
This study result demonstrated that DMSB was capable of discriminating parallel G4s from other DNA motifs based on the dual-site simultaneously binding mode. The recognizing signatures of parallel G4s presented here were very simple by using only UV-vis study. This dual-site binding mode of DMSB also exhibited higher specificity of parallel G4 motifs than that of one endstacking binding mode probes. All these findings might provide opportunities in designing high specificity G4 probes.
Acknowledgments
This present study was supported by Major National Basic Research Projects (973,No. 2013CB733701),National Natural Science Foundation of China (Nos. 81072576,91027033, 21302188,21205121,21305145 and 31200576),and Chinese Academy of Sciences (No. KJCX2-EW-N06-01).
| [1] | A. Siddiqui-Jain, C.L. Grand, D.J. Bearss, L.H. Hurley, Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription, Proc. Natl. Acad. Sci. U. S. A. 99 (2002) 11593–11598. |
| [2] | J.T. Davis, G-quartets 40 years later: from 50-GMP to molecular biology and supramolecular chemistry, Angew. Chem. Int. Ed. 43 (2004) 668–698. |
| [3] | S. Burge, G.N. Parkinson, P. Hazel, A.K. Todd, S. Neidle, Quadruplex DNA: sequence,topology and structure, Nucleic Acids Res. 34 (2006) 5402–5415. |
| [4] | J.R. Williamson, G-quartet structures in telomeric DNA, Annu. Rev. Biophys.Biomol. Struct. 23 (1994) 703–730. |
| [5] | J.R. Williamson, M.K. Raghuraman, T.R. Cech, Monovalent cation-induced structure of telomeric DNA: the G-quartet model, Cell 59 (1989) 871–880. |
| [6] | J.L. Huppert, S. Balasubramanian, G-quadruplexes in promoters throughout the human genome, Nucleic Acids Res. 35 (2007) 406–413. |
| [7] | G.W. Collie, G.N. Parkinson, The application of DNA and RNA G-quadruplexes to therapeutic medicines, Chem. Soc. Rev. 40 (2011) 5867–5892. |
| [8] | Q.F. Yang, J.F. Xiang, S. Yang, et al., Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: I. Recognizing mixed G-quadruplex in human telomeres, Chem. Commun. ([2TD$DIF]2009) 1103–1105. |
| [9] | Q.F. Yang, J.F. Xiang, S. Yang, et al., Verification of specific G-quadruplex structure by using a novel cyanine dye supramolecular assembly: II. The binding characterization with specific intramolecular G-quadruplex and the recognizing mechanism,Nucleic Acids Res. 38 (2010) 1022–1033. |
| [10] | L.J. Yu, Q.F. Yang, J.F. Xiang, et al. Analyst (2015), http://dx.doi.org/10.1039/c4an01912a. |
| [11] | Q.F. Yang, J.F. Xiang, S. Yang, et al., Verification of intramolecular hybrid/parallel G-quadruplex structure under physiological conditions using novel cyanine dye H-aggregates: both in solution and on Au film, Anal. Chem. 82 (2010) 9135–9137. |
| [12] | B. Pagano, A. Virno, C.A.Mattia, et al., Targeting DNA quadruplexeswith distamycin A and its derivatives: an ITC and NMR study, Biochimie 90 (2008) 1224–1232. |
| [13] | C. Rajput, R. Rutkaite, L. Swanson, I. Haq, J.A. Thomas, Dinuclear monointercalating RuII complexes that display high affinity binding to duplex and quadruplex DNA, Chemistry 12 (2006) 4611–4619. |
| [14] | T. Wilson, M.P. Williamson, J.A. Thomas, Differentiating quadruplexes: binding preferences of a luminescent dinuclear ruthenium(II) complex with four-stranded DNA structures, Org. Biomol. Chem. 8 (2010) 2617–2621. |
| [15] | Z. Li, J.H. Tan, J.H. He, et al., Disubstituted quinazoline derivatives as a new type of highly selective ligands for telomeric G-quadruplex DNA, Eur. J. Med. Chem. 47(2012) 299–311. |
| [16] | P.[1TD$DIF]Z. Zhang, H.[1TD$DIF]L. Yang, C.[1TD$DIF]C. Li, et al., Synthesis of novel, azasugar-modified anthraquinone derivatives and their cytotoxicity, Chin. Chem. Lett. 25 (2014)1057–1059. |
| [17] | E.W. White, F. Tanious, M.A. Ismail, et al., Structure-specific recognition of quadruplex DNA by organic cations: influence of shape, substituents and charge,Biophys. Chem. 126 (2007) 140–153. |
| [18] | S. Cosconati, L. Marinelli, R. Trotta, et al., Structural and conformational requisites in DNA quadruplex groove binding: another piece to the puzzle, J. Am. Chem. Soc.132 (2010) 6425–6433. |
| [19] | W. Gai, Q.F. Yang, J.F. Xiang, et al., A dual-site simultaneous binding mode in the interaction between parallel-stranded G-quadruplex [d(TGGGGT)]4 and cyanine dye 2,20-diethyl-9-methyl-selenacarbocyanine bromide, Nucleic Acids Res. 41(2013) 2709–2722. |
| [20] | F.M. Hamer, The Chemistry of Heterocyclic Compounds, Interscience, New York,1964. |
| [21] | L.G.S. Brooker, F.L. White, Studies in the cyanine dye series: I. A new method of preparing certain carbocyanines, J. Am. Chem. Soc. 57 (1935) 547–551. |
| [22] | A.H. Herz, Aggregation of sensitizing dyes in solution and their adsorption onto silver halides, Adv. Colloid Interface Sci. 8 (1977) 237–298. |
| [23] | A.H. Herz, Dye–dye interactions of cyanines in solution and at AgBr surfaces,Photogr. Sci. Eng. 18 (1974) 323–335. |
| [24] | W. West, S. Pearce, Dimeric state of cyanine dyes, J. Phys. Chem. 69 (1965)1894–1903. |
| [25] | E.G. Mcrae, M. Kasha, Enhancement of phosphorescence ability upon aggregation of dye molecules, J. Chem. Phys. 28 (1958) 721–722. |
| [26] | A.K. Chibisov, H. Go¨ rner, Photophysics of aggregated 9-methylthiacarbocyanine bound to polyanions, Chem. Phys. Lett. 357 (2002) 434–439. |
| [27] | J.T. McPhee, E. Scott, N.E. Levinger, A. Van Orden, Cy3 in AOT reverse micelles: I.Dimer formation revealed through steady-state and time-resolved spectroscopy,J. Phys. Chem. B 115 (2011) 9576–9584. |
| [28] | G. Laughlan, A.I. Murchie, D.G. Norman, et al., The high-resolution crystal structure of a parallel-stranded guanine tetraplex, Science 265 (1994)520–524. |
| [29] | G.N. Parkinson, M.P.H. Lee, S. Neidle, Crystal structure of parallel quadruplexes from human telomeric DNA, Nature 417 (2002) 876–880. |
| [30] | S. Neidle, S. Burge, G.N. Parkinson, P. Hazel, A.K. Todd, Quadruplex DNA: sequence,topology and structure, Nucleic Acids Res. 34 (2006) 5402–5415. |
| [31] | J.W. Yan, W.J. Ye, S.B. Chen, et al., Development of a universal colorimetric indicator for G-quadruplex structures by the fusion of thiazole orange and isaindigotone skeleton, Anal. Chem. 84 (2012) 6288–6292. |




