b Shanghai Collaborative Innovation Center for Biomanufacturing Technology, Shanghai 200237, China
Diamides,the newest major class of insecticides,aroused interests worldwide and played key roles in Integrated Pest Management (IPM) for their broad insecticidal spectra,high efficiency,low mammalian toxicity,and low residual [1]. The first member of diamides was the phthalamide flubendiamide, which was discovered by the Japanese company Nihon Nohyaku [2, 3] and jointly developed with Bayer Crop Sciences. Then,the second one,chlorantraniliprole,was discovered by Dupont with an anthranilic diamide structure [4, 5]. As a fasting growing class of insecticides in modern crop protection,anthranilic diamides attracted more considerable attention [6]. In recent years,based on the structure of chlorantraniliprole,lots of anthranilic compounds with different level of insecticidal activities were synthesized and investigated by many groups [7, 8, 9, 10, 11, 12, 13, 14, 15].
Though many investigations consistent with the hypothesis that chlorantraniliprole act on the insect ryanodine receptor (RyR) [16, 17, 18],very few was known about its binding mode,as well as the bioactive conformation,which deeply restricted the further discovery of newanthranilic diamide. The bioactive conformation of a drug is the special one adapted in ligand-receptor interaction and plays guiding roles in new biochemical molecular design [19]. Generally,the bioactive conformation of a ligand could be identified by ligand-proteincrystal structure ormoleculardockingstudies. The ryanodine receptor is a complicated tetrameric transmembrane Ca2+ channel [20, 21]. Unfortunately,it presents major challenges to current experimental techniques,because of its enormous size,its integral membrane protein nature,and dynamic features [22]. To date,no crystal structure of the ion channel has been reported, though some mammal subunits have been resolved [23, 24]. It is impossible to capture the binding pocket of anthranilic diamides, and even the bioactive conformation.
Liu et al. [25] reported 3D-QSAR models of a series of anthranilic diamides,of which cis-amide conformations were adopted,thus the bioactive conformation of anthranilic diamides was still unclear. In the bioactive conformation research of herbicides, Zhang et al. [26] successfully predicted the bioactive conformations of Protoporphyrinogen oxidase inhibitors by combining DFTbased potential energy scanning with QSAR analysis,which provided a good computational approach for bioactive conformation exploration. CoMFA is a very popular QSAR method,which has been reported highly sensitive to the different space orientations of compounds [27]. It was generally considered that QSAR models based on the bioactive conformation should result in much better statistical significance than that of non-bioactive conformations. Thus,the shortage of CoMFA also provided an useful approach to identify the bioactive conformation of a compound from a series of low energy conformations.
DFT methods have been successfully used in preparing the structures of compounds [28, 29, 30]. In the present study,DFT/B3LYP based potential energy (PES) scanning was carried out to explore the low-energy conformations of chlorantraniliprole. Then,based on the low-energy conformations,the structure alignments were performed for a series of anthranilic diamide compounds, respectively,followed by detailed CoMFA and CoMSIA analyses. The results revealed the bioactive conformation of chlorantraniliprole, which might provide some useful clues for future design of new anthranilic diamide insecticides.
2. Experimental 2.1. Potential energy surface scanning and low-energy conformationsChlorantraniliprole (Fig. 1A) was selected for potential energy surface scanning to obtain low-energy conformations. For the convenience of calculation,a simplified structure model (Fig. 1C) was used in the PES scanning,of which the chloropyridine moiety was replaced with a hydrogen atom to remove its bulky effects. Then,the PES scanning was performed by B3LYP/6-31G(d,p) method,based on the two rotatable bonds including C5-C9 and C4-C13 with a step of 30°. All minimum conformations of modelM obtained by the PES scanning were optimized at the B3LYP/6- 31G(d,p) level. Finally,the chlorantraniliprole geometry were reconstructed by adding chloropyridine moiety and optimized by B3LYP/6-31G(d,p) method to find the corresponding low-energy conformations. All PES scanning and structure optimizations were performed using Gaussian 09 software (Gaussian,Inc.,Wallingford CT,2009).

|
Download:
|
| Fig. 1.The structures of chlorantraniliprole (A,top view; B,front view) and scanning model (C). | |
Data on series of anthranilic diamides and their insecticidal activities were retrieved from two literatures of the same group [31, 32]. Compounds were tested against the diamondback moth (plutella xylostel,Px) under the same assay procedures. Reported LC50 values were converted into the corresponding pLC50 (-log LC50) and listed with all the structures in Table 1. Twentyone compounds were divided into a training set comprising 16 compounds,and a test set comprising five compounds selected randomly for CoMFA and CoMSIA studies.
|
|
Table 1 Structures and insecticidal activity values of compounds. |
The conformations of compounds were modified from chlorantraniliprole and optimized by B3LYP/6-31G(d,p) method. The optimized structures of all compounds were imported into SYBYL, among which the most active compound 5 was selected as a template for superimposition. Then,CoMFA analysis was applied to explore the bioactive conformation of chlorantraniliprole. All parameters used for CoMFA modeling were remained their default values. With standard options for scaling of variables,the regression analysis was carried out using the full cross-validated partial least square (PLS) leave-one-out (LOO) method to get the q2 value. The final model (non-cross-validated conventional analysis) was developed with the optimum number of components to yield the r2 value.
CoMSIA models were also evaluated by using the standard settings. The default value of 0.3 was used as attenuation factor. The statistical evaluation for the CoMSIA analyses was performed in the same way as described for CoMFA. Finally,the model quality was evaluated in terms of the LOO cross-validated correlation coefficient (q2),the non-cross-validation correlation coefficient (r2),the standard error of estimate (SEE),and the F-statistic for analysis (F).
3. Results and discussion3.1. Low-energy conformations of chlorantraniliprole
The bioactive conformation of a compound is not always the lowest-energy one,but must be one with low energy [26]. Potential energy surface scanning analysis was performed to find out all of the possible low-energy conformations. Fig. 2 depicted the PES scanning results for model M. The minimums in the surface (Fig. 2B) were selected and optimized by B3LYP/6-31G(d,p) method. Finally,10 minimum conformations were obtained,of which the lowest four (M1-M4) were showed in Fig. 3. Conformations M1 and M2 have the same energy values and depicted the lowest energy conformations. Conformations M3 and M4 also showed equal energy values,which were higher than M1 and M2 with △E of 5.24 kcal/mol. Thus,only conformations M1 and M2 were remained for further analysis. The transfer energy barrier between M1 and M2 calculated at B3LYP/6-31G(d,p) level was 15.8 kcal/mol in vacuum,revealed that M1 and M2 could not be converted to each other easily.

|
Download:
|
| Fig. 2.Potential energy surface scanning results for model M. (A) front view,(B) upward view (the points marked with * were selected for further optimization). X axis for dihedral angle C4-C5-C9-O12 and Y axis for dihedral angle C3-C4-N13-C14. | |

|
Download:
|
| Fig. 3.Four low energy conformations and relative energies of model M. | |
Based on conformations M1 and M2,the low energy conformations of chlorantraniliprole were constructed by adding the chloropyridine moiety. Considering two possible orientations of Cl atomin the chloropyridine group,four low-energy conformations (CI-CIV) of chlorantraniliprole were finally optimized by B3LYP/6- 31G(d,p) method (Fig. 4). Frequency calculation at the same level revealed that they were all minimums in potential energy surface. The relative energies of the four conformations (CI-CIV) were very similar,ranging from 0 to 0.45 kcal/mol,in which an intramolecular N-H…O H-bond could be observed.

|
Download:
|
| Fig. 4.Four local energy minima conformations of chlorantraniliprole and their relative energies. | |
CoMFA method is sensitive to the conformation of compounds, and even small conformation changes could generate dramatic influence on the results ofCoMFA analysis. It is generally considered thatQSARmodels based on the bioactive conformation should result in much better statistical significance than that of non-bioactive conformations. The results of CoMFA statistical analysis were presented in Table 2. For the CoMFA models,the conventional noncross- validated coefficient r2 values were 0.921,0.915,0.909,and 0.977,respectively for the overlapped structures based on four conformations marked CI,CII, CIII,and CIV. The corresponding cross-validated coefficient q2 values of 0.419,0.369,0.343,and 0.740 were obtained with optimum number of components (ONC) 3, respectively. The SEE values were 0.153,0.158,0.164,and 0.083,the F values were 46.672,43.008,39.863,and 168.197,respectively. The highest q2,r2,and F values,as well as the lowest SEE value,indicated that the CoMFA model based on conformation CIV was clearly better than the others. The statistics of CoMFA analysis (Table 2,Table S1, and Fig. S2 in Supporting information) implied that CIV might be the bioactive conformation of chlorantraniliprole. For clearly show, conformation CIV were also depicted in Fig. 1A and B. For CIV,if the phenyl was regarded as the paper plane and the methyl group connected the benzene ring was pointing to the down direction,the O and H atoms forming the N-H…O H-bond were out the phenyl plane,and the pyridinewas also pointing to the downdirection with the substituted Cl inside the phenyl plane.
|
|
Table 2 Statistical results for CoMFA and CoMSIA models. |
CoMSIA analyses were also performed to validate whether CIV is the bioactive conformation. CoMSIA models were carried out using steric,electrostatic,hydrophobic,H-bond donor,and H-bond acceptor fields in different combinations. The statistics of CoMSIA analyses were also presented in Table 2. The PLS analysis of CoMSIA models generated from the superimposed structures based on four conformations marked CI,CII,CIII,and CIV showed q2 values of 0.146,-0.413,-0.118,and 0.628,respectively. The corresponding r2 values as 0.854,0.734,0.785,and 0.952, respectively. The SEE values were 0.208,0.280,0.252,and 0.123, the F values were 23.344,11.017,14.635,and 78.970,respectively. The conformation CIV showed also the best q2,r2,SEE,and F values in all CoMSIA models (Table 2,Table S1,and Fig. S3 in Supporting information). Both CoMFA and CoMSIA analysis indicated that CIV should be the bioactive conformation of chlorantraniliprole.
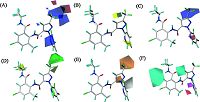
3.3. 3D-QSAR contour map analysesCoMFA and CoMSIA contour maps for compounds based on the conformation CIV were showed in Fig. 5. To aid visualization, compound 5 was displayed as the reference molecule. The CoMFA electrostatic contour map was shown in Fig. 5A. The blue and red contours depicted the regions of desirable positive and negative electrostatic interactions. A large blue contour surrounding the 3- position of the pyrazole ring indicated that negative charge was associated with reduced activity. For example,the activities of compounds 9 and 10 with a Cl atom at the 3-position of the pyrazole ring was less than that of compounds 7 and 8 with a Br atomat the sameposition.Andamediumblue contourwas observed at the 6-position of the chloropyridine ring,whichwas in agreement with that the activity of compound 1 (benzene ring)was higher than compound 4 (pyridine ring). The CoMFA steric contour map was depicted in Fig. 5B. The beneficial and adverse steric interactions were displayed ingreenand yellowcontours,respectively.However, no useful information could be obtained in Fig. 5B.
|
Download:
|
| Fig. 5.Contour maps with compound 5. (A) CoMFA electrostatic field. (B) CoMFA steric field. (C) CoMSIA electrostatic field. (D) CoMSIA steric field. (E) CoMSIA hydrophobic field. (F) CoMSIA H-bond donor and acceptor fields. | |
In the case of CoMSIA,additional insights from hydrophobic,Hbond donor and H-bond acceptor features were got,of which the contour maps were also depicted in Fig. 5. The CoMSIA electrostatic contour map was shown in Fig. 5C. Similar to the results from CoMFA,a large blue contour was also found around the 3-positon of the pyrazole ring. From the steric field,a large green contour was near the 3-position of the pyrazole ring (Fig. 5D). It agreed with the results that compounds 17 and 18 substituted with -OCH3 group at the 3-position of pyrazole ring,showed lower activities than compounds 20 and 21 with -OCH2CF3 group.
The CoMSIA hydrophobic contour map was showed in Fig. 5E. The favorable hydrophobic field was showed in orange contours, and the favored hydrophilic field was showed as white contours. A large hydrophobic region presented at 3-positon of the pyrazole ring,which indicated that increasing hydrophobicity in this area could improve activities. It was consistent with the findings that compound 6 with a -CF3 group at the 3-position of the pyrazole ring was higher activities than compound 17 with a -OCH3 group at the same position.
The CoMSIA H-bond donor and acceptor contour map was showed in Fig. 5F. The favorable areas of H-bond donor and acceptor contours were displayed with cyan and magenta,and the unfavorable regions were showed by purple and red,respectively. Two cyan contours showed two H-bond donors favored regions near the amide groups connected with the phenyl ring,which indicated that the two amide groups connected with the phenyl ring are very important for high insecticidal activity.
4. ConclusionIn summary,by combining DFT-based potential energy surface scanning with CoMFA and CoMSIA analyses,the bioactive conformations of anthranilic diamide insecticides were disclosed from a series of low energy conformations. An intramolecular N- H…O H-bond was found important for maintaining the bioactive conformation of chlorantraniliprole. For the bioactive conformation, if the phenyl was regarded as the paper plane and the methyl group connected with phenyl was pointing to the down direction, the O and H atoms forming the N-H…O H-bond were out the phenyl plane,and the pyridine was also pointing to the down direction with the substituted Cl inside the phenyl plane. The findings might provide some clues for future insecticide design. In addition,it also indicated that DFT-based potential energy surface scanning combined with CoMFA analysis was a good approach for exploring the bioactive conformation of a compound,when the target structure was unknown.
AcknowledgmentThe authors thanks for the financial support from the National Key Technology R&D Program of China (No. 2011BAE06B05).
Appendix A. Supplementary dataSupplementary data associated with this article can be found,in the online version,at http://dx.doi.org/10.1016/j.cclet.2015.04.010.
| [1] | A. Jeanguenat, The story of a new insecticidal chemistry class: the diamides, Pest Manag. Sci. 69 (2013) 7-14. |
| [2] | T. Masaki, N. Yasokawa, S. Fujioka, et al., Quantitative relationship between insecticidal activity and Ca2+ pump stimulation by flubendiamide and its related compounds, J. Pestic. Sci. 34 (2009) 37-42. |
| [3] | M. Tohnishi, H. Nakao, T. Furuya, et al., Flubendiamide, a novel insecticide highly active against lepidopterous insect pests, J. Pestic. Sci 30 (2005) 354-360. |
| [4] | A. Dinter, K.E. Brugger, N.M. Frost, M.D. Woodward, Chlorantraniliprole (Rynaxypyr): a novel DuPontTM insecticide with low toxicity and low risk for honey bees (Apis mellifera) and bumble bees (Bombus terre stris) providing excellent tools for uses in integrated pest management, Julius-Kühn-Archiv 423 (2010) 84-96. |
| [5] | D. Cordova, E.A. Benner, M.D. Sacher, et al., Anthranilic diamides: a new class of insecticides with a novel mode of action, ryanodine receptor activation, Pestic. Biochem. Physiol. 84 (2006) 196-214. |
| [6] | D.B. Sattelle, D. Cordova, T.R. Cheek, Insect ryanodine receptors: molecular targets for novel pest control chemicals, Invert. Neurosci. 8 (2008) 107-119. |
| [7] | Y.B. Chen, J.L. Li, X.S. Shao, X.Y. Xu, Z. Li, Design, synthesis and insecticidal activity of novel anthranilic diamides with benzyl sulfide scaffold, Chin. Chem. Lett. 24 (2013) 673-676. |
| [8] | D.A. Clark, G.P. Lahm, B.K. Smith, J.D. Barry, D.G. Clagg, Synthesis of insecticidal fluorinated anthranilic diamides, Bioorg. Med. Chem. 16 (2008) 3163-3170. |
| [9] | M. Luo, Q.C. Chen, J. Wang, et al., Novel chlorantraniliprole derivatives as potential insecticides and probe to chlorantraniliprole binding site on ryanodine receptor, Bioorg. Med. Chem. Lett. 24 (2014) 1987-1992. |
| [10] | J.J. Ou, X.K. Zhu, L. Wang, et al., Synthesis and bioactivity study of 2-acylaminosubstituted N0-benzylbenzohydrazide derivatives, J. Agric. Food Chem. 60 (2012) 10942-10951. |
| [11] | T.P. Selby, G.P. Lahm, T.M. Stevenson, et al., Discovery of cyantraniliprole, a potent and selective anthranilic diamide ryanodine receptor activator with cross-spectrum insecticidal activity, Bioorg. Med. Chem. Lett. 23 (2013) 6341-6345. |
| [12] | B.L. Wang, H.W. Zhu, Y. Ma, et al., Synthesis, insecticidal activities, and SAR studies of novel pyridylpyrazole acid derivatives based on amide bridge modification of anthranilic diamide insecticides, J. Agric. Food Chem. 61 (2013) 5483-5493. |
| [13] | J. Wu, B.A. Song, D.Y. Hu, M. Yue, S. Yang, Design, synthesis and insecticidal activities of novel pyrazole amides containing hydrazone substructures, Pest Manag. Sci. 68 (2012) 801-810. |
| [14] | J.F. Zhang, J.Y. Xu, B.L. Wang, et al., Synthesis and insecticidal activities of novel anthranilic diamides containing acylthiourea and acylurea, J. Agric. Food Chem. 60 (2012) 7565-7572. |
| [15] | Y.Y. Zhou, Q. Feng, F.J. Di, et al., Synthesis and insecticidal activities of 2,3-dihydroquinazolin-4(1H)-one derivatives targeting calcium channel, Bioorg. Med. Chem. 21 (2013) 4968-4975. |
| [16] | A.K. Isaacs, S.Z. Qi, R. Sarpong, J.E. Casida, Insect ryanodine receptor: distinct but coupled insecticide binding sites for [N-C3H3]chlorantraniliprole, flubendiamide, and [3H]ryanodine, Chem. Res. Toxicol. 25 (2012) 1571-1573. |
| [17] | G.P. Lahm, D. Cordova, J.D. Barry, New and selective ryanodine receptor activators for insect control, Bioorg. Med. Chem. 17 (2009) 4127-4133. |
| [18] | S.Z. Qi, J.E. Casida, Species differences in chlorantraniliprole and flubendiamide insecticide binding sites in the ryanodine receptor, Pestic. Biochem. Physiol. 107 (2013) 321-326. |
| [19] | S.R. LaPlante, H. Nar, C.T. Lemke, et al., Ligand bioactive conformation plays a critical role in the design of drugs that target the hepatitis C virus NS3 protease, J. Med. Chem. 57 (2014) 1777-1789. |
| [20] | I.I. Serysheva, E.V. Orlova, W. Chiu, et al., Electron cryomicroscopy and angular reconstitution used to visualize the skeletal muscle calcium release channel, Nat. Struct. Biol. 2 (1995) 18-24. |
| [21] | M. Radermacher, V. Rao, R. Grassucci, et al., Cryo-electron microscopy and threedimensional reconstruction of the calcium release channel/ryanodine receptor from skeletal muscle, J. Cell Biol. 127 (1994) 411-423. |
| [22] | L. Kimlicka, F. Van Petegem, The structural biology of ryanodine receptors, Sci. China Life Sci. 54 (2011) 712-724. |
| [23] | F. Van Petegem, Ryanodine receptors: structure and function, J. Biol. Chem. 287 (2012) 31624-31632. |
| [24] | L.N. Sun, L. Cui, C.H. Rui, et al., Modulation of the expression of ryanodine receptor mRNA from Plutella xylostella as a result of diamide insecticide application, Gene 511 (2012) 265-273. |
| [25] | G.Y. Liu, X.L. Ju, J. Cheng, Z.Q. Liu, 3D-QSAR studies of insecticidal anthranilic diamides as ryanodine receptor activators using CoMFA, CoMSIA and DISCOtech, Chemosphere 78 (2010) 300-306. |
| [26] | L. Zhang, G.F. Hao, Y. Tan, et al., Bioactive conformation analysis of cyclic imides as protoporphyrinogen oxidase inhibitor by combining DFT calculations, QSAR and molecular dynamic simulations, Bioorg. Med. Chem. 17 (2009) 4935-4942. |
| [27] | J.W. Zou, C.C. Luo, H.X. Zhang, et al., Three-dimensional QSAR of HPPD inhibitors, PSA inhibitors, and anxiolytic agents: effect of tautomerism on the CoMFA models, J. Mol. Graph. Model. 26 (2007) 494-504. |
| [28] | T.H. Li, X.G. Xie, G.B. Du, A theoretical study on the water-mediated asynchronous addition between urea and formaldehyde, Chin. Chem. Lett. 24 (2013) 85-88. |
| [29] | A.A. Peyghan, M.T. Baei, S. Hashemian, M. Moghimi, Adsorption of nitrous oxide on the (6,0) magnesium oxide nanotube, Chin. Chem. Lett. 23 (2012) 1275-1278. |
| [30] | S. Xia, Y. Feng, J.G. Cheng, et al., QAAR exploration on pesticides with high solubility: an investigation on sulfonylurea herbicide dimers formed through π-π stacking interactions, Chin. Chem. Lett. 25 (2014) 973-977. |
| [31] | G.P. Lahm, T.M. Stevenson, T.P. Selby, et al., RynaxypyrTM: a new insecticidal anthranilic diamide that acts as a potent and selective ryanodine receptor activator, Bioorg. Med. Chem. Lett. 17 (2007) 6274-6279. |
| [32] | G.P. Lahm, T.P. Selby, J.H. Freudenberger, et al., Insecticidal anthranilic diamides: a new class of potent ryanodine receptor activators, Bioorg. Med. Chem. Lett. 15 (2005) 4898-4906. |