Chinese prescription is the fundamental method of traditional Chinese clinical medication and therefore the essence of traditional Chinese medicine (TCM). In modern science,the chemical components of Chinese prescriptions are responsible for their medicinal effects. Therefore,identifying the chemical components of Chinese prescriptions is important to understand their functional mechanisms. However,Chinese prescriptions contain complicated components,and identification and analysis of these multiple components can be difficult. With the development of modern analytical techniques,a variety of tandem high-resolution mass spectrometers (HRMS),such as Q-TOF,IT-TOF,LTQ-FTICRMS, and LTQ-Orbitrap,have been widely used for the detection and identification of compounds in TCM [1, 2, 3, 4]. With advantages such as excellent resolution,high mass accuracy,superior sensitivity, fast scan rate,and multiple-stage mass spectrometry scanning (MSn),these mass spectrometers can provide HRMS and MSn data for nearly all compounds present in TCM using a simple data acquisition method [5, 6]. However,automatically acquired mass spectrometric data contain numerous endogenous components. Some challenges that need to be addressed include discovering relevant chemical components from the data automatically collected by the equipment and selecting a method that quickly identifies the structures of the components using the obtained mass spectrometric data.
Recently,mass spectral tree similarity has been introduced and applied to the identification of unknown compounds based on a library of known compounds [7, 8, 9, 10]. The reported methods [7, 8, 9, 10] of mass spectral tree similarity have presented practical values for the identification of chemical compounds,especially for the identification of metabolites in metabolomics. In our previous study [11],a novel mass spectral tree similarity filter (MTSF) technique was developed to discover and identify related metabolites based on the similarity of high resolution and multiple-stage mass spectrometric data between the detected compounds and templated library compounds.
This work is the first to apply MTSF technique to study the chemical components of the active fraction from ‘‘Xiao-Xu-Ming decoction (XXMD)’’. XXMD was a traditional Chinese prescription and first recorded in ‘‘Bei Ji Qian Jin Yao Fang’’. The formula consists of 12 crude drugs including Saposhnikovia divaricata (Turcz.) Schischk.,Scutellaria baicalensis Georgi,Paeonia lactiflora Pall., Glycyrrhiza uralensis Fisch.,Zingiber officinale Rosc.,Stephania tetrandra S. Moore,Panax ginseng C. A. Mey.,Cinnamomum cassia Presl,Prunus armeniaca L. var. ansu Maxim.,Ephedra sinica Staph, Ligusticum chuanxiong Hort.,and Aconitum carmichaeli Debx. in a ratio of 3:3:3:3:3:3:6:6:6:9:9:9 on a dry weight basis. It has been widely used to treat theoplegia and the effects of theoplegia in China. Through this study,we increased the application range of the MTSF technique and obtained satisfactory results. 2. Experimental 2.1. Materials
Paeoniflorin (R1),prim-O-glucosylcimifugin (R2),cimifugin (R4),40-O-b-D-glucosyl-5-O-methylvisamminol (R5),baicalin (R6), baicalein (R9),glycyrrhizic acid (R10),wogonin (R11),chrysin (R12),and glycyrrhetinic acid (R14) were purchased from the National Institute for Control of Pharmaceutical and Biological Products (Beijing,China). Liquiritigenin (R3) was obtained from the Dalian Fusheng Pharmaceutical Co.,Ltd. Wogonoside (R8) was ordered from the Shanghai Usea Biotech Co.,Ltd. Oroxylin A-7-Oglucuronide (R7) and oroxylin A (R13) were from Baoji Herbest Bio-Tech Co.,Ltd. Acetonitrile and methanol of LC/MS reagent grade was obtained from Mallinckrodt Baker,Inc. (Phillipsburg,NJ, USA). Purified water was generated by using a Millipore water purification system (Millipore,Billerica,MA,USA). Analytical grade formic acid was obtained from Merck Inc. (Darmstadt,Germany). The active fraction of XXMD was screened by Wang et al. [12] and shown to exert a similar pharmacological effect to that of XXMD in treating theoplegia and its effects. 2.2. Chromatography and mass spectrometry conditions
Separation was carried out on a Surveyor LC Plus system equipped with a Surveyor MS Pump Plus,a Surveyor Autosampler, and a Surveyor PDA Plus detector. Samples were separated on a Thermo BDS HYPERSIL C18 column (150 mm × 2.1 mm,3 µm) protected by an Agilent SB-C18 guard column (12.5 mm × 4.6 mm, 5 µm). The mobile phase consisted of 0.1% (v/v) formic acid in water (A) and acetonitrile (B) delivered at a flow rate of 0.2 mL min-1 using a gradient program as follows: beginning with 5% B and maintaining for 5 min,then reaching 30% B at 25 min,reaching 40% B at 35 min,reaching 80% B at 45 min,and maintaining for 5 min. The system then returned to the initial conditions within 60 s,and the column was reconditioned for 9 min. Column temperature was maintained at 30 ℃,and the sample injection volume was 5 µL. UV detection was set at 270 nm and 254 nm separately.
A Thermo Scientific LTQ FT was connected to the Thermo Scientific Surveyor LC Plus system via an electrospray ionization (ESI) interface. Ultrahigh-purity helium (He) was used as the collision gas,and high-purity nitrogen (N2) was used as the nebulizing gas. The operating parameters in the positive ion mode were as follows: ion spray voltage at 4.0 kV,capillary temperature at 250 ℃,capillary voltage at 46 V,sheath gas flow rate of 40 (arbitrary units),auxiliary gas flow rate of 10 (arbitrary units), sweep gas flow rate of 3 (arbitrary units),and tube lens at 90 V. Compounds were detected by full-scan mass analysis from m/z 100 to 1200 at a resolving power of 50,000 with data-dependent MS2 analysis triggered by the two most abundant ions from full-scan mass analysis,followed by MS3 analysis of the most abundant product ions. Collision-induced dissociation (CID) was performed with an isolation width of 2 Da. The collision energy was set to 35%. Dynamic exclusion was conducted by utilizing a repeat count of one,prior to exclusion. Each mass-to-charge value resided on the dynamic exclusion list for 30 s after the data-dependent MS2 experiment. Upon data-dependent analyses incorporating dynamic exclusion,high mass-to-charge accuracy and multiple-stage mass spectrometry of compounds in samples could be acquired automatically. Data acquisition was performed with Xcalibur version 2.0 SR2 software (Thermo Fisher Scientific,San Jose,CA, USA). 2.3. Sample preparation
A 50 mg sample of the active fraction of XXMT was accurately weighed,transferred to a 10 mL volumetric flask with 9 mL methanol,vortexed for 30 s,ultrasonicated for 3 min,and then diluted with methanol to obtain the test solution with the concentration at 5 mg/mL. The test solution was filtered through a 0.45 mm nylon filter film before analysis. 2.4. MTSF technique workflow
HRMS and multiple-stage mass spectrometric data of all detected compounds were obtained and changed to mass spectral tree data by Mass Frontier version 7.0 software (Thermo Fisher Scientific). Then,the mass spectral tree data of the fourteen reference compounds (R1-R14) which had been identified in our previous studies [13] were imported into a new established library as templated compounds. The mass frontier software was used to calculate the similarity of mass spectral tree data of compounds detected in the active fraction of XXMT with those contained in the library,which is expressed by match similarity score [10, 14]. The highest matching score is 1000,while the no matching score is 0. The higher the similarity score,the more likely it was that the compound resembled library compounds,and score 1000 indicates the detected compound is the library compound. According to our research experience [11],using scores over 200 as a limit can effectively filter interference endogenous compounds and fish out the related compounds in the library. Finally,the fished compounds were identified by further using the high accurate molecular mass combined with multiple-stage mass spectrometric data and fragmentation rules. 3. Results and discussion
Optimized chromatographic conditions were adjusted based on the previous fingerprint analysis method [13]. Such adjustments included a change to a small size of the column (250 mm × 4.6 mm,5 µm changed to 150 mm × 2.1 mm,3 µm) and optimization of the elution gradient of the mobile phase. A suitable simple data acquisition method was mainly developed through the maximum development of LTQ FT acquisition capability. By optimizing injection time (200 ms for FT,50 ms for LTQ),micro scan (1),and resolution (50,000),the scan rate reached more than 0.5 Hz. Data dependent scanning was combined with the dynamic exclusion technique to collect multiple-stage mass spectrometry data automatically. The conditions of the datadependent scanning were determined from the multiple-stage mass spectrometry data based on the two most abundant ions from full-scan mass analysis,followed by MS3 analysis of the most abundant product ions. To collect the multiple-stage mass spectrometry data rapidly,the dynamic exclusion technique was used,which sets up a dynamic depot to exclude all collected parent ions. The main setting parameters include the repeat count,repeat duration,exclusion list size,and exclusion duration. Through optimization,several parameters were set as follows: repeat count,1; repeat duration,10 s; exclusion list size,50; and exclusion duration,30 s. This data acquisition method was used to ensure that the multiple-stage mass spectrometry data of at least the top 30 strongest ions will be collected within the chromatographic peak width time of 30-60 s to meet the test requirements.
Active fraction of XXMD analysis was performed using the above high performance liquid chromatography-high resolution mass spectrometry (HPLC-HRMS) method with the automatically collected mass spectral data. In one injection,the HRMS and multiple-stage mass spectrometric data of nearly all compounds present in the active fraction of XXMD were obtained. However, when the instrument was set to collect the mass spectrometric data automatically,the spectra contained a large amount of irrelevant ion mass spectrometric information. Establishing a means by which comprehensive compounds can be successfully distinguished during the post-acquisition data processing procedure has became a significant issue for the investigation of chemical components of Chinese prescriptions by tandem HRMS.
In this study,the MTSF technique,which was first developed as a means to rapidly discover comprehensive metabolites from multiple active components in a complicated biological matrix, was used to discover and identify the chemical components of XXMD. HRMS and multiple-stage mass spectrometric data were converted into mass spectra trees by a total extraction component detection algorithm,and a total of 3362 compounds were automatically detected. The mass spectral trees of 14 templated compounds were imported into a newly-built material user library for component discovery. The statistical significance of the scores was calculated by comparing the mass spectral trees data of compounds detected in XXMT with 14 templated compounds in the library. A threshold for match similarity scores was set by manually checking each compound in the previous study,and a score of above 200 could effectively filter out interference compounds [11]. For this study,less than 400 of these compounds had match similarity scores above 200. In other words,using this limit,the MTSF technique could eliminate almost 90% of interference compounds. Finally,after manual examining accurate mass,fragmentation patterns,multiple-stage mass data,and retention time of these 400 compounds,we identified 50 related chemical components (Fig. 1).

|
Download:
|
| Fig. 1.Using MTSF technique,50 related chemical components (red indication,1-50) of Xiao-Xu-Ming decoction were discovered and identified base on the 14 templated compounds (green target recognition,R1-R14). In addition,4 unrelated compounds with high abundance were identified manually (orange indication,N1-N4). | |
In addition,the mass spectrometry tree diagram that can be established by using this technology enables the comparison of visualized compounds,which is convenient for structure identification. An example of identification of compounds in the active fraction of XXMD using MTSF technique is shown in Fig. 2. These results show that this technology has the advantage of rapidly identifying components of Chinese prescription (for the representative detail mass data of related compounds fished by MTSF technique see Table 1). Notably,the unrelated peak in Fig. 1 is the compounds whose structures exhibited significant difference from the 14 known compounds. Identification of them could be carried out by manually elucidation,and in this experiment,an attempt was made to interpret some high abundance compounds (such as N1-N4). Take N1 as example,the proposed compound N1 had a protonated molecular mass of m/z 458.166,and its proposed formula was C20H28NO11 with an error of 0.68 ppm. Its multiple stage mass spectra gave ions of [M + H-162]+ at m/z 296 and [M + H-324]+ at m/z 134. This formula was searched in the Chemspider database and the structure was identified as amygdalin. This compound was obtained from Prunus armeniaca L. var. ansu Maxim.,which is a component herb of XXMD [15]. The other three unrelated compounds were also identified by this method. According to the above results,the MTSF technological process can rapidly discover and identify related compounds through some templated compounds. In outlook on the application of this technology to the research of phytochemistry,besides simplification in the processes of quick identification of chemical compounds,because of the relevance it has made between the components of phytomedicine and templated compounds,we can make out the compounds that we are interested in or exclude those that we do not desire in accordance with our research objectives.

|
Download:
|
| Fig. 2.An example of identification of compounds in the active fraction of XXMD using MTSF technique. Because 7-O-glucoside-baicalein (compound 22) had the same product ions as baicalin (one templated compound),it can be effectively filtered from endogenous interference compounds by MTSF. | |
| Table 1 Representative detail mass data of related compounds fished by MTSF technique (using R6 and R9 as templated compounds). |
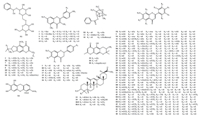
A total of 68 compounds were identified in the active fraction of XXMD. However,in our previous study [13],we just identified 14 major compounds of XXMT using manual mining approach. Briefly, the results demonstrated that MTSF technique should be useful to rapid discovery and identification of compounds in Chinese prescription. Aside from showing the advantages of this technology in terms of rapid discovery and identification of the chemical components of Chinese prescription,this study also proved that MTSF can be applied to the targeted phytochemical separation (Fig. 3).
|
Download:
|
| Fig. 3.The structures of identified 68 compounds in the active fraction of XXMD. The numbers assigned to templated compounds are R1-R14,assigned to related compounds are 1-50,assigned to unrelated compounds are N1-N4. | |
The authors would like to thank the Natural Science Foundation of Beijing (No. 7133252) for financial support of this work.
| [1] | H. Wang, G. Yan, A. Zhang, et al., Rapid discovery and global characterization of chemical constituents and rats metabolites of Phellodendri amurensis cortex by ultra-performance liquid chromatography-electrospray ionization/quadrupoletime-of-flight mass spectrometry coupled with pattern recognition approach, Analyst 138 (2013) 3303-3312. |
| [2] | Y. Liang, H. Hao, A. Kang, et al., Qualitative and quantitative determination of complicated herbal components by liquid chromatography hybrid ion trap time-of-flight mass spectrometry and a relative exposure approach to herbal pharmacokinetics independent of standards, J. Chromatogr. A 1217 (2010) 4971-4979. |
| [3] | Y. Dai, F.J. Tu, Z.H. Yao, et al., Rapid identification of chemical constituents in traditional Chinese medicine Fufang preparation Xianling Gubao capsule by LClinear ion trap/orbitrap mass spectrometry, Am. J. Chin. Med. 41 (2013) 1181-1198. |
| [4] | Y. Rao, M. McCooeye, Z. Mester, Mapping of sulfur metabolic pathway by LC orbitrap mass spectrometry, Anal. Chim. Acta 721 (2012) 129-136. |
| [5] | M. Paul, J. Ippisch, C. Herrmann, et al., Analysis of new designer drugs and common drugs of abuse in urine by a combined targeted and untargeted LC-HR-QTOFMS approach, Anal. Bioanal. Chem. 406 (2014) 4425-4441. |
| [6] | X.C. Zhu, Y.P. Chen, R. Subramanian, Comparison of information dependent acquisition, SWATH, and MSAll techniques in metabolite identification study employing UHPLC-Q-TOF mass spectrometry, Anal. Chem. 86 (2014) 1202-1209. |
| [7] | M. Rojas-Cherto, J.E. Peironcely, P.T. Kasper, et al., Metabolite identification using automated comparison of high-resolution multistage mass spectral trees, Anal. Chem. 84 (2012) 5524-5534. |
| [8] | J.E. Peironcely, M. Rojas Chertó, A. Tas, et al., Automated pipeline for de novo metabolite identification using mass-spectrometry-based metabolomics, Anal. Chem. 85 (2013) 3576-3583. |
| [9] | P.T. Kasper, M. Rojas-Chertó, R. Mistrik, et al., Fragmentation trees for the structural characterisation of metabolites, Rapid Commun. Mass Spectrom. 26 (2012) 2275-2286. |
| [10] | M.T. Sheldon, R. Mistrik, T.R. Croley, Determination of ion structures in structurally related compounds using precursor ion fingerprinting, J. Am. Soc. Mass Spectrom. 20 (2009) 370-376. |
| [11] | Y. Jin, C.S. Wu, J.L. Zhang, Y.F. Li, A new strategy for the discovery of epimedium metabolites using high-performance liquid chromatography with high resolution mass spectrometry, Anal. Chim. Acta 768 (2013) 111-117. |
| [12] | Y. Wang, H. Qin, X. He, G. Du, Activity evaluation of components and preparation of effective components group of Xiaoxuming decoction for anti-cerebral ischemic, China J. Chin. Mater. Med. 36 (2011) 2140-2144. |
| [13] | Y. Wang, C. Ding, K. Du, et al., Identification of active compounds and their metabolites by high-performance liquid chromatography/electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry from Xiao-Xu-Ming decoction (XXMD), Rapid Commun. Mass Spectrom. 23 (2009) 2724-2732. |
| [14] | P. Sander, Substance identification of ion trap MS/MS spectra in a MS/MS library, in: Proceedings of 47th ASMS Conference on Mass Spectrometry and Allied Topics, Dallas, TX, 1999. |
| [15] | Q. Lv, L.Z. Yi, H.Y. Yi, et al., Chromatographic fingerprint of Semen Armeniacae Amarae based on high-performance liquid chromatogram and chemometric methods, Anal. Methods 1 (2012) 299-308. |





