b Key Laboratory for Advanced Materials & Department of Chemistry, East China University of Science and Technology, Shanghai 200237, China;
c School of Information Science and Engineering, East China University of Science and Technology, Shanghai 200237, China
The nanopore has become a unique tool for single molecule detection with the advantages of label free and high-throughput sensing [1, 2]. It has been successfully used to explore a large variety of biomolecules including oligonucleotides [3, 4, 5, 6, 7],peptides [11, 12, 13],proteins [8, 9, 10],and protein-DNA complexes [14, 15]. Previous studies have showed that a biological nanopore is an ultra-sensitive sensor for monitoring the conformational changes of biomolecule which is induced by the biological weak interactions [16, 17, 18, 19, 20]. In particular,nanopore technique offers the prospect of sequencing a human genome at the expense of -$1000 within 24 h [21, 22, 23]. Generally,in a nanopore experiment, an individual molecule is driven into a nanopore under a biased voltage in the electrolyte solution. When the molecule transverses through the nanopore,it will produce a characteristic ionic blockage. After statistical analysis of currents and durations of the blockages,the properties of an individual molecule,such as the composition,length and secondary structure,could be elucidated. During the translocation process of biomolecules at a certain electrophoretic force,the blockage duration not only depends on the size of the biomolecule but also the interactions between nanopore and the analyte. Therefore,the histogram for the blockage duration appears to be a half Gaussian and half exponential distribution [24]. The curve follows a relatively steep rise and fall,but the blockage durations larger than the value of Gaussian peak is approximated by an exponential decay. In a conventional data analysis process,the distributions of durations are fitted by exponential functions. However,the exponential fittings barely cover the whole distribution of durations,which exclude all of the events with the durations shorter than the value of Gaussian peak. The inappropriateness of exponential distributions only reveals the random walks of the biomolecules translocation rather than other deterministic phenomena. Therefore,the exponential function is insufficient to model the blockage durations of translocation events in nanopore analysis.
Exponentially modified Gaussian function could be used to describe process which begins with Gaussian distribution and ends with exponential distribution [25]. It has been widely applied to analyze peaks in chromatography [26],model the distributions of intermitotic time and extracts variabilities of protein expression in biology [27]. Herein,EMG was introduced into analyzing the blockage durations for nanopore-based single molecule detection. The EMG was compared to conventional methods and validated by analyzing the stimulated distributions as well as the histograms for oligonucleotides translocations by nanopore experiments. Our results demonstrated that,compared with traditional exponential function,EMG provided a better fit and covered the entire range of the distributions. In particular,the fitted parameters of EMG could be directly used to discriminate the sequence length of oligonucleotides at single molecule level. 2. Experimental
α-Hemolysin (α-HL) was purchased from Sigma-Aldrich (St. Louis,MO,USA) and used without purification. Diphytanoylphosphatidyl-choline was purchased from Avanti Polar Lipids Inc. (Alabaster,AL,USA). Poly(dA)60 (oligo 1) and the sequence 5'-ACCTGGGGGAGTATTGTAAAAAAGAATGTCGCAAAAAAAAAAA-3'(oligo 2) were synthesized by Invitrogen Life Technologies (Shanghai,China). The final concentration of the analyte in 1 mL cischamber was 0.15 umol/L. Prior to use,DNA solutions were annealed at 95°C for 10 min and then cooled to room temperature. Millipore water (18 MVcm-1) was used throughout the experiments. Unless otherwise noted,all other chemicals were of analytical grade.
The nanopore electrical recording was conducted according to our previous studies [15, 16, 17, 28, 29]. The lipid bilayers were created by applying diphytanoyl-phosphatidyl-choline (30 mg/mL) in decane (≥99%,Sigma-Aldrich,St. Louis,MO,USA) to a 150mm orifice in a 1 mL bilayer cup integrated into a lipid bilayer chamber (Warner Instruments,Hamden,CT,USA) filled with KCl (1.0 mol/L) and Tris-HCl (10 mmol/L,pH 7.8). Temperature was fixed at 25±0.5°C by amounting the lipid bilayer chamber on a thermal stage (Dagan Corporation,Minneapolis,MN,USA). The a-HL was injected adjacent to the aperture in the cis chamber,and pore insertion was determined by a well-defined jump in current value. Then,oligo 1 and oligo 2 were added in thecissolution under a voltage of +100 mV,respectively.
The ionic currents were acquired using a ChemClamp amplifier (Dagan Corporation,Minneapolis,MN,USA),filtered with a 3-pole low-pass Bessel filter at 3 kHz and sampled at 100 kHz by DigiData 1440A/D converter (Axon Instruments,Forest City,CA,USA). The ionic currents were recorded by a PC running PClamp 10.2 (Axon Instruments,Forest City,CA,USA). The recorded current data were analyzed by a home-made matlab program.
Theoretical simulation: A one-dimensional lattice Monte Carlo algorithm was used to simulate the translocation process of singlestrand DNA through nanopore whose cross-section was close to the pore size of α-HL. The detailed simulation process was described in a previous study [30]. The blockage durations of each simulation were recorded and sorted into histograms. 3. Results and discussion
As a probability distribution,the EMG function is the combination of normal and exponential random variables. The equivalent form of the EMG could be written as follows:
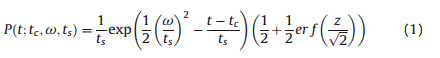
 )is the error
function evaluated at ƒ=(
)is the error
function evaluated at ƒ=( ),in which z=((t-tc)/ω)-(ω/ts).
),in which z=((t-tc)/ω)-(ω/ts).
As shown in Fig. 1,we generated examples of EMG function through a Matlab program,with appropriate domain ranging from 0 to 10. Obviously,the EMG showed a characteristic positive skew, which is consistent with the histogram of duration of translocation events in nanopore. In addition,position,width and skew of the peak are tunable by adjusting the parametersv,tc and ts . Thus, EMG is able to cover the whole distribution of blockage durations for nanopore experiments.

|
Download:
|
| Fig. 1. The three parameters tc,ts,and vcould influence the shape of EMG in peak position,width and skewness. | |
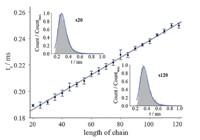
Since the shape of the duration histogram originates from the translocation behavior of each oligonucleotide,it cannot be known ab initio. Here,we use the Monte Carlo simulation to generate the translocation events of oligonucleotides for the purpose of validating the introduced EMG. In the Monte Carlo simulation, the nanopore was simplified to a one-dimension channel as long as 40 bases. The oligonucleotides were considered as ideal,selfavoiding,and rod-like chains with lattice coordination number set to 3. The simulation was carried out at atmospheric temperature of 288.15 K and external electric field was 2. Chains with length from 20 to 120 bases were applied to simulate the translocation processes,respectively. For each chain,we generated 100,000 events for the substantial statistic analysis. Then,the histograms of duration time were fitted by both EMG and exponential function. The expression of the exponential function applied in this paper was ƒ(x)=λ1 exp(-λ2x),where λ1 and λ2 were both parameters of the function. The bin-width of the duration histograms was set to 0.01 ms,while the histograms were normalized by dividing the count number of each bin by the maximum count number among all bins. Fig. 2 illustrates the EMG fitting examples of oligonucleotides with 20 and 120 bases,respectively. The parameters (tc ,ts,ω) were obtained by fitting the EMG function to the histogram through trust region algorithm in Matlab program. The commonly used goodness of fit criteria (R-squared, R2 ) was applied to check the fitting performances. Compared with the traditional exponential fittings,the values of R2 of all fitted results carried out by EMG were much higher (Table 1). Therefore,the histograms of blockage durations were well fitted by the EMG. The fitted value tc,which affects the peak value of distribution,exhibited a linear dependence of the chain lengths. As the length of oligonucleotides increased from 20 to 120 bases,the value of tclinearly increased from 0.19 to 0.25 (Fig. 2). In fact,relationships between the chain length and the peak position in the histogram have been systemically investigated in quite a lot previous literatures [24, 31, 32]. Since the parameter tc in EMG function affects the position of peak,chain length is reasonable to exhibit a linear relationship with the value of parameter tc. Therefore,the fitting results of EMG could be used to discriminate the length of oligonucleotides.
| Table 1 The fitted results of EMG for the simulated events of s20,s45,s70,s95 and s120.a |
|
Download:
|
| Fig. 2. The values oftc as a function of chain length ranging from 20 to 120 bases. Insertion: Duration histogram of the simulated events ofs20ands120,which were fitted by EMG distribution respectively. | |
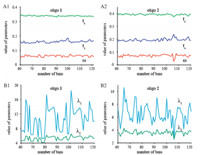
To examine the performance of EMG in fitting the experimental data,we applied it to fit the blockage durations recorded froma-HL nanopore experiments of Poly(dA)60 (oligo 1) and a random sequence (oligo 2),respectively. The numbers of events used in the statistic analyzing are 809 and 668 events for oligo1and oligo 2, respectively. As shown in Fig. 3,the curves of EMG are well matched with the edge of both the duration histograms of oligo1 and oligo 2. As a result,the R2 of EMG fitting was 0.97 for oligo 1. For the random sequences oligo 2,the value of R2 was 0.80,which is higher than the traditional exponential fitting of 0.65 and 0.45, respectively. In the traditional exponential fitting,the varying binwidth of a histogram highly affects the fitted values of blockage durations. Here,we investigated the influence of bin-setting on the fitted values of EMG. The bin-width was set from 0.075 to 0.0375 ms with an interval of -6.25×10-4ms. The values of EMG parameters (tc ,ts,ω) were hardly influenced by the setting of binwidth (Fig. 4A) compared with the values of Exponential parameters (λ1,λ2) in Fig. 4B. Table 2 lists the standard deviations of the fitted parameters. EMG fitting shows that the discretions of the parameters from the average values are lower than 1%,while exponential fittings gave larger standard deviations of more than 30%,even to 300%. As confirmed by standard deviation,there were no remarkable changes of the fitted parameters carried out by the EMG fittings among the wide range of bin-width. Therefore,the fitted results of EMG are stable and reliable for the nanopore analysis.

|
Download:
|
| Fig. 3. Duration histograms of oligo1(A) and oligo2(B) fitted by EMG (blue curve). (For interpretation of the references to color in this figure legend,the reader is referred to the web version of this article.) | |
| Table 2 Mean value and standard deviations of the fitted parameters for oligo 1 and oligo 2 with bin-width increased from 0.075 to 0.0375 ms at an interval of -6.25×10-4ms. |
|
Download:
|
| Fig. 4. The values of parameters in EMG function (A) and exponential function (B) for fitting the duration histogram of oligo1and oligo2with the bin-width ranging from 0.075 to 0.0375 ms,corresponding to the number of bins from 60 to 120 in the figure. | |
Our results demonstrated a well performed EMG function for the nanopore statistic analysis at single molecule level. EMG significantly improves the stability and reliability of the data analysis through the wide range of bin-width. Compared with exponential function in traditional nanopore analysis,EMG offers a more accurate description of duration histogram for both real and simulated translocations. More importantly,the value of fittedtc depends on the length of oligonucleotides,which could be further used to analyze the single-molecule behaviors of oligonucleotides. Hence,the introduced EMG functions provide a better description for the statistic nanopore analysis and will facilitate the nanopore based single-molecule detection. Acknowledgments
The authors acknowledge funding of the National Natural Science Foundation of China (No. 21327807). Y.-T. Long is grateful for funds from the National Science Fund for Distinguished Young Scholars of China (No. 21125522). Y.-L. Ying thanks the Sino-UK Higher Education Research Partnership for PhD Studies.
| [1] | J. Kasianowicz, E. Brandin, D. Branton, D. Deamer, Characterization of individual polynucleotide molecules using a membrane channel, Proc. Natl. Acad. Sci. U.S.A. 93 (1996) 13770-13773. |
| [2] | Y.L. Ying, J. Zhang, R. Gao, Y.T. Long, Nanopore-based sequencing and detection of nucleic acids, Angew. Chem. Int. Ed. 52 (2013) 13154-13161. |
| [3] | M. Akeson, D. Branton, J.J. Kasianowicz, E. Brandin, D. Deamer, Microsecond timescale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules, Biophys. J. 77 (1999) 3227-3233. |
| [4] | N. An, A.M. Fleming, H.S. White, C.J. Burrows, Crown ether-electrolyte interactions permit nanopore detection of individual DNA abasic sites in single molecules, Proc. Natl. Acad. Sci. U.S.A. 109 (2012) 11504-11509. |
| [5] | Y. Wang, D. Zheng, Q. Tan, M.X. Wang, L.Q. Gu, Nanopore-based detection of circulating microRNAs in lung cancer patients, Nat. Nanotechnol. 6 (2011) 668-674. |
| [6] | S. Liu, B. Lu, Q. Zhao, et al., Boron nitride nanopores: highly sensitive DNA singlemolecule detectors, Adv. Mater. 25 (2013) 4549-4554. |
| [7] | S. Wen, T. Zeng, L. Liu, et al., Highly sensitive and selective DNA-based detection of mercury(II) with alpha-hemolysin nanopore, J. Am. Chem. Soc. 133 (2011) 18312-18317. |
| [8] | J. Sha, T. Hasan, S. Milana, et al., Nanotubes complexed with DNA and proteins for resistive-pulse sensing, ACS Nano 7 (2013) 8857-8869. |
| [9] | J. Nivala, D.B. Marks, M. Akeson, Unfoldase-mediated protein translocation through an [alpha]-hemolysin nanopore, Nat. Biotechnol. 31 (2013) 247-250. |
| [10] | D. Rotem, L. Jayasinghe, M. Salichou, H. Bayley, Protein detection by nanopores equipped with aptamers, J. Am. Chem. Soc. 134 (2012) 2781-2787. |
| [11] | T.C. Sutherland, Y.T. Long, R.I. Stefureac, et al., Structure of peptides investigated by nanopore analysis, Nano Lett. 4 (2004) 1273-1277. |
| [12] | L. Movileanu, J.P. Schmittschmitt, J. Martin Scholtz, H. Bayley, Interactions of peptides with a protein pore, Biophys. J. 89 (2005) 1030-1045. |
| [13] | H.Y. Wang, Y.L. Ying, Y. Li, Y.T. Long, Peering into biological nanopore: a practical technology to single-molecule analysis, Chem. -Asian J. 5 (2010) 1952-1961. |
| [14] | F. Olasagasti, K.R. Lieberman, S. Benner, et al., Replication of individual DNA molecules under electronic control using a protein nanopore, Nat. Nanotechnol. 5 (2010) 798-806. |
| [15] | Y. Ying, X. Zhang, Y. Liu, et al., Single molecule study of the weak biological interactions between P53 and DNA, Acta Chim. Sin. 71 (2013) 44-50. |
| [16] | Y.L. Ying, D.W. Li, Y. Li, J.S. Lee, Y.T. Long, Enhanced translocation of poly(dt) 45 through an α-hemolysin nanopore by binding with antibody, Chem. Commun. 47 (2011) 5690-5692. |
| [17] | Y.L. Ying, H.Y. Wang, T.C. Sutherland, Y.T. Long, Monitoring of an ATP-binding aptamer and its conformational changes using an alpha-hemolysin nanopore, Small 7 (2011) 87-94. |
| [18] | Y.L. Ying, D.W. Li, Y. Liu, et al., Recognizing the translocation signals of individual peptide-oligonucleotide conjugates using an α-hemolysin nanopore, Chem. Commun. 48 (2012) 8784-8786. |
| [19] | H.Y. Wang, Z. Gu, C. Cao, J. Wang, Y.T. Long, Analysis of a single alpha-synuclein fibrillation by the interaction with a protein nanopore, Anal. Chem. 85 (2013) 8254-8261. |
| [20] | Y.L. Ying, J. Zhang, F.N. Meng, et al., A stimuli-responsive nanopore based on a photoresponsive host-guest system, Sci. Rep. (2014), http://dx.doi.org/10.1038/ srep01662. |
| [21] | D. Branton, D.W. Deamer, A. Marziali, et al., The potential and challenges of nanopore sequencing, Nat. Biotechnol. 26 (2008) 1146-1153. |
| [22] | B. Gyarfas, F. Olasagasti, S. Benner, et al., Mapping the position of DNA polymerase-bound DNA templates in a nanopore at 5Åresolution, ACS Nano 3 (2009) 1457-1466. |
| [23] | K.R. Lieberman, G.M. Cherf, M.J. Doody, et al., Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase, J. Am. Chem. Soc. 132 (2010) 17961-17972. |
| [24] | A. Meller, L. Nivon, E. Brandin, J. Golovchenko, D. Branton, Rapid nanopore discrimination between single polynucleotide molecules, Proc. Natl. Acad. Sci. U.S.A. 97 (2000) 1079-1084. |
| [25] | D. Hanggi, P.W. Carr, Errors in exponentially modified Gaussian equations in the literature, Anal. Chem. 57 (1985) 2394-2395. |
| [26] | X. Li, V.L. McGuffin, Theoretical evaluation of methods for extracting retention factors and kinetic rate constants in liquid chromatography, J. Chromatogr. A 1203 (2008) 67-80. |
| [27] | A. Golubev, Exponentially modified Gaussian (EMG) relevance to distributions related to cell proliferation and differentiation, J. Theor. Biol. 262 (2010) 257-266. |
| [28] | H.Y. Wang, Y.L. Ying, Y. Li, H.B. Kraatz, Y.T. Long, nanopore analysis of amyloid peptide aggregation transition induced by small molecules, Anal. Chem. 83 (2011) 1746-1752. |
| [29] | Y. Liu, Y.L. Ying, H.Y. Wang, et al., Real-time monitoring of the oxidative response of a membrane-channel biomimetic system to free radicals, Chem. Commun. 49 (2013) 6584-6586. |
| [30] | M.G. Gauthier, G.W. Slater, A Monte Carlo algorithm to study polymer translocation through nanopores. I. Theory and numerical approach, J. Phys.Chem. 128 (2008) 065103. |
| [31] | A. Meller, D. Branton, Single molecule measurements of DNA transport through a nanopore, Electrophoresis 23 (2002) 2583-2591. |
| [32] | J. Li, M. Gershow, D. Stein, E. Brandin, J. Golovchenko, DNA molecules and configurations in a solid-state nanopore microscope, Nat. Mater. 2 (2003) 611-615. |