b College of Chemistry and Molecular Engineering, Peking University, Beijing 100871, China;
c Jozef Stefan Institute, Jamova 39, SI-1000 Ljubljana, Slovenia
1. Introduction
Quantitative analysis of DNA at low concentration is very important in many fields,including forensic medicine,gene engineering,pharmaceuticals,clinical medicine,and so on. Various detection approaches have been reported,such as surface plasmon (SP) resonance [1, 2, 3],resonance light scattering (RLS) [4],fluorimetry [5],chemiluminescence [6, 7, 8],SNPs [9], electrochemistry [10, 11, 12, 13, 14, 15],and UV-vis spectrophotometry [16- 21]. Among them,optical and electrochemical methods have drawn great attention. Most reported spectrophotometric methods were based on the decrease of absorbance due to the binding reaction of DNA with color reagents,resulting in limited sensitivity.
In recent years,nanomaterials used in detection of biomolecules have attracted much attention due to their high sensitivity and selectivity [22, 23]. Various nanomaterials such as nanoparticles, nanotubes,and nanosheets have been synthesized and employed in detection of biological recognition events [24]. The results indicate that application of nanomaterials have brought about a great improvement in both the sensitivity and selectivity of biosensing devices. Conversely,these have accelerated synthesis and preparation of new nanomaterials.
Molybdenum-chalcogenide-halide nanowires (NWs) which are composed of molybdenum (Mo),sulfur (S) and iodine (I) in the form Mo6S9-xIx (MoSI) are a new class of quasi-onedimensional objects,having two different stoichiometries:Mo6S3I6 andMo6S4.5I4.5. An identical skeletal structure is composed of indistinguishable one dimensional polymer chains of molybdenum- sulfur-iodine clusters,strongly joined together by anions (either S or I). The individual nanowires are joined together into bundles by weak Van der Waals forces. The materials have strong anisotropy,large Young moduli along the wires,very small shear moduli,and controllable electronic properties. Compared to carbon nanotubes,MoSI NWs have some significant advantages such as straight forward synthesis,monodisperse diameters,and metallic properties [25, 26, 27, 28, 29, 30]. Our preliminary work has demonstrated a novel electrochemical sensor for highly sensitive detection of natural double-stranded deoxyribonucleic acid (dsDNA) based on thionin (Th) attached to MoSI NWs selfassembled on a gold electrode [10].
Since UV absorbance is used to determine concentration of nucleic acids,based on our previous work on amplified optical detection of proteins derived from MoSI NWs [31],herein,we demonstrate that the MoSI NWs can serve as an excellent signalintensifying nanomaterial for highly sensitive and label-free detection of DNA by UV spectrophotometry. 2. Experimental 2.1. Reagents
MoSI NWs were fabricated by direct synthesis from elemental material that had been mixed in the desired stoichiometries,as described elsewhere [25]. Powders composed of aggregates of individual nanowires were obtained with two different stoichiometries Mo6S3I6 and Mo6S4.5I4.5. The stoichiometries were determined by a combination of techniques such as chemical analysis, high-resolution transmission electron microscopy,and X-ray photoelectron spectroscopy (XPS),as reported in the literature [30]. In this work,all studies were carried out on Mo6S3I6 NWs. Fish sperm DNA was obtained from Acros; fish sperm DNA solution in 0.20 MNaCl,gave a ratio of UV absorbance at 260 nm and 280 nm, A260/A280,of ca. 1.85,indicating that DNA was sufficiently free of protein and RNA [32]. The concentration of fish sperm DNA standard stock solution (1.223 μg/mL) was spectrophotometrically determined at 260 nm. All other reagents were of analytical grade. Water was triply distilled with a quartz apparatus.
In 2-propanol,1.00 μg/mL MoSI NWs dispersions were prepared. The dispersions were initially sonicated for 2 min using a high-power ultrasonic tip (120 W,60 kHz) followed by a mild sonication for 2 h using a low power ultrasonic bath. Prior to use, the dispersions were resonicated for 20 min to obtain uniform suspension. 2.2. Apparatus
UV-vis absorption spectra were recorded on a U-4100 UV-vis- NIR spectrophotometer (Hitach,Japan) in a 1.0 cm quartz cuvette within a wavelength range from 200 nm to 400 nm.
Circular dichroism (CD) spectra were obtained by a J-720 JASCO spectropolarimeter (JASCO,Japan) at room temperature,and a rectangular quartz cell of 1 cm path length was used to obtain spectra from 210 nm to 350 nm by taking points every 0.5 nm.
Transmission electron microscopic (TEM) characterizations were obtained by JEM-2100 (JEOL,Japan) with a working voltage of 200 kV. The samples were prepared as followed: DNA solute, MoSI NWs solute and their mixed solution in isopropanol were dropped on holeycarbon film 300-mesh copper grids,respectively, and then allowed to dry in air. 3. Results and discussion
Fig. 1 showed the characteristic extinction spectra of natural DNA at 260 nm in the absence and presence of MoSI NWs solute. With the addition of different volumes of the MoSI NW solutes in the range from 0.10 mL to 1.00 mL in isopropanol into 3.40 μg/mL DNA solution while keeping the total volume of all the solutions the same at 3.00 mL,the extinction at 260 nm was greatly enhanced with increasing amounts of the MoSI NWs (curves c,d and e),whereas both the MoSI NWs and DNA had weak UV absorption(curves a and b) at 260 nm. This revealed that MoSI NWs act as a signal amplifying reagent for the UV response of DNA. The possible reason for this is interaction between DNA and the MoSI NWs,resulting in greater DNA coverage on the NWs surface,and further enhancing the extinction signal,which is clearly observed by the TEM images in Fig. 2. The appearance of MoSI NWs was thin and clear-shaped (Fig. 2A),and it became thick and rough after immersion into the dsDNA solution (Fig. 2C),which evidenced the immobilization of DNA on MoSI NWs. The extinction of DNA-MoSI NWs exhibited a reduced behavior (Fig. 1,curve f) when the amount of the MoSI NW solutes was 1.50 mL. We believe that excessive MoSI NWs encourages large DNA-MoSI NWs aggregates, resulting in a reduced absorbance.

|
Download:
|
| Fig. 1.UV–vis spectra of MoSI NWs solute (a) and 3.40 μg/mL DNA with addition of different volumes of MoSI NWs solute: 0 mL (b), 0.10 mL (c), 0.50 mL (d), 1.00 mL (e), 1.50 mL (f) in a fixed volume (3.00 mL) of isopropanol. | |

|
Download:
|
| Fig. 2.TEM images of MoSI NWs (A), DNA (B) and MoSI NWs–DNA ensemble (C). The sample (C) was prepared as follows: MoSI NWs solute was mixed with 2.43 μg/mL DNA, and then dropped on a holeycarbon film of 300-mesh copper grid and allowed to dry in air. | |
When changing the concentration of DNA in a 1.00 mL of MoSI NWs isopropanol solution,the spectra were recorded as shown in Fig. 3A,and the concentration dependence of the extinction fits well to linear increase between 0.0289 μg/mL and 11.68 μg/mL. The linear regression equations are: A = 0.1064 + 0.1639 c (μg/mL) (R = 0.997,n = 8) and A = -0.07159 + 0.3343 c (μg/mL) (R = 0.995, n = 17) with the concentration of DNA in ranges of 0.0289- 0.549 μg/mL (Fig. 3B) and 0.549-11.68 μg/mL (Fig. 3C),respectively. The real determination limit is 28.9 ng/mL (Fig. 3A inset).
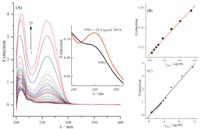
|
Download:
|
| Fig. 3.(A): UV extinction spectra of 1.00 mL of MoSI NWs solution with addition of different concentrations of DNA (μg/mL): 0 (1), 0.0289 (2), 0.0578 (3), 0.0867 (4), 0.116 (5), 0.173 (6), 0.289 (7), 0.405 (8), 0.549 (9), 0.694 (10), 0.838 (11),0.983 (12), 1.273 (13),1.56 (14),1.85 (15), 2.14 (16), 2.43 (17), 2.72 (18), 3.01 (19), 3.29 (20), 3.87 (21), 4.45 (22), 5.90 (23),8.79 (24) and 11.68 (25). Inset shows a real determination limit of 28.9 ng/mL. (B) and (C): the working curves between extinction and the concentration of DNA. | |
Circular dichroism (CD) spectroscopy is widely used for characterizing changes in DNA conformation when DNA interacts with other compounds [33, 34, 35, 36, 37]. In order to further verify the interaction between MoSI NWs and DNA,CD spectra were also conducted. Fig. 4 shows the CD spectra of natural fish sperm DNA, MoSI NWs,and their mixture,respectively. The CD spectrum of DNA (Fig. 4) exhibits a positive band at 273 nmand a negative band at 246 nm,which are characteristics of B-form DNA,the most frequently observed conformation of DNA [38]. After incubation with MoSI NWs,the CD amplitudes of DNA reduce,which reveals the conformational transformation from a B-form duplex to a hairpin transition [38]. This provides a support for the interaction between DNA and MoSI NWs,similar to the interaction between DNA and H2Ti3O7 nanotubes in the reported literature [39]. We speculate that the interaction might originate from hydrogen bonds involving sulfur atoms and N-H groups based on the reported literatures [40, 41, 42],bringing about the binding of DNA to MoSI NWs. These illustrate that the enhancement of UV extinction signal is caused by the greater DNA coverage on the MoSI NW surface and the conformational transformation of DNA.

|
Download:
|
| Fig. 4.Circular dichroism spectra of DNA solution in the absence and presence of MoSI NWs: 2.17 μg/mL DNA (a), 2.17 μg/mL DNA + 1.00 mL NWs (b), 1.00 mL NWs (c). | |
In conclusion,we have demonstrated that MoSI NWs are a promising nanostructure material for the development of highly sensitive biosensors. The binding of natural DNA to MoSI NWs enables the amplified detection of DNA using UV-vis spectrophotometry. The optical biosensor is based on one soluble phase of MoSI NWs in isopropanol and binding of DNA to them. Enhancement of the extinction for DNA in the presence of MoSI NWs has enabled a simple DNA detection scheme to be developed. The new label free MoSI NWs-derived amplification bioassay is expected to open new opportunities for DNA analysis and biosensor construction.
AcknowledgmentThis project was supported by the National Natural Science Foundation of China (No. 20875005).
| [1] | T. Endo, K. Kerman, N. Nagatani, Y. Takamura, E. Tamiya, Label-free detection of peptide nucleic acid-DNA hybridization using localized surface plasmon resonance based optical biosensor, Anal. Chem.77 (2005) 6976-6984. |
| [2] | D. Pollard-Knight, E. Hawkins, D. Yeung, et al., Immunoassays and nucleic-acid detection with a biosensor based on surface-plasmon resonance, Ann. Bid. Clin.48 (1990) 642-646. |
| [3] | R. Karlsson, A. Michaelsson, L. Mattsson, Kinetic-analysis of monoclonal antibodyantigen interactions with a new biosensor based analytical system, J. Immunol. Methods 145 (1991) 229-240. |
| [4] | S. Feng, Z.P. Li, S.H. Zhang, Z. Fang, Recent advance of resonance light scattering technique for the determination of nucleic acids, Spectrosc. Spect. Anal.24 (2004) 1676-1680. |
| [5] | L. Li, J.H. Yang, X. Wu, C.X. Sun, G.J.Zhou, Fluorimetric determination of nucleic acid using the enhancement of terium-gadolinium-nucleic acid-cetylpyridine bromide system, Talanta 59 (2003) 81-87. |
| [6] | Y.J. Tang, Z.Y. Li, N.Y. He, et al., Preparation of functional magnetic nanoparticles mediated with PEG-4000 and application in pseudomonas aeruginosa rapid detection, J. Biomed. Nanotechnol.9 (2013) 312-317. |
| [7] | F. Wang, C. Ma, X. Zeng, et al., Chemiluminescence molecular detection of sequence-specific HBV-DNA using magnetic nanoparticles, J. Biomed. Nanotechnol. 8 (2012) 786-790. |
| [8] | N. He, F. Wang, C. Ma, et al., Chemiluminescence analysis for HBV-DNA hybridization detection with magnetic nanoparticles based DNA extraction from positive whole blood samples, J. Biomed. Nanotechnol. 9 (2013) 267-273. |
| [9] | S. Li, H. Liu, Y. Jia, et al., A novel SNPs detection method based on gold magnetic nanoparticles array and single base extension, Theranostics 2 (2012) 967-975. |
| [10] | H. Lin, H.M. Cheng, L. Liu, et al., Thionin attached to a gold electrode modified with self-assembly of Mo6S9-xIx nanowires for amplified electrochemical detection of natural DNA, Biosen. Bioelectron. 26 (2011) 1866-1870. |
| [11] | K.Q. Deng, C.X. Li, Y.L. Ling, G.R. Xu, X.F. Li, Fabrication of poly(2,6-pyridinedicarboxylic acid)/MWNTs modified electrode for simultaneous determination of guanine and adenine in DNA, Chin. Chem. Lett.22 (2011) 981-984. |
| [12] | W. Zhang, T. Yang, D.M. Huang, K. Jiao, Electrochemical sensing of DNA immobilization and hybridization based on carbon nanotubes/nano zinc oxide/chitosan composite film, Chin. Chem. Lett.19 (2008) 589-591. |
| [13] | J.Wang, Nanomaterial-based electrochemical biosensors, Analyst 130 (2005) 421-426. |
| [14] | A. Erdem, D.O. Ariksoysal, H. Karadeniz, et al., Electrochemical genomagnetic assay for the detection of hepatitis B virus DNA in polymerase chain reaction amplicons by using disposable sensor technology, Electrochem. Commun. 7 (2005) 815-820. |
| [15] | S.J. Park, T.A. Taton, C.A.Mirkin, Array-based electrical detection of DNA with nanoparticle probes, Science 295 (2002) 1503-1506. |
| [16] | A.A. Killeen, A visible spectrophotometric assay for submicrogram quantities of DNA including PCR-amplified DNA, Microchem. J. 52 (1995) 333-340. |
| [17] | C.Z. Huang, K.A. Li, S.Y. Tong, Spectrophotometry of nucleic acids by their effect on the complex of cobalt(Ⅱ) with 4-[(5-chloro-2-pyridyl)azo]-1,3-diaminobenzene, Anal. Chim. Acta 345 (1997) 235-242. |
| [18] | Y.M. Hao, H.X. Shen, Spectrophotometric determination of nucleic acids using palladium(Ⅱ) complex with 2-(5-bromo-2-pyridylazo)-5-diethylaminophenol, Anal. Chim. Acta 413 (2000) 87-94. |
| [19] | W.H. Si, Y.Q. Zi, Y.F. Tu, Spectrophotometric determination of deoxyribonucleic acid by its quenching effect on acridine orange, Spectrosc. Spect. Anal.28 (2008) 412-414. |
| [20] | H. Wang, W.R. Li, Y. Lu, N.N. Fu, H.S. Zhang, Spectrophotometric determination of DNA using a near infrared probe 1,10-disulfobutyl-3,3,30,30-tetramethylindotricarbocyanine, Spectrochim. Acta A 61 (2005) 2103-2107. |
| [21] | T.J. Li, H.X. Shen, Y.J. Luo, Spectrophotometric determination of deoxyribonucleic acid labeling with ethyl violet, Chin. J. Anal. Chem.26 (1998) 1372-1375. |
| [22] | J.J. Storhoff, S.S. Marla, P. Bao, et al., Gold nanoparticle-based detection of genomic DNA targets on microarrays using a novel optical detection system, Biosen. Bioelectron.19 (2004) 875-883. |
| [23] | Q.C. Zou, Q.J. Yun, G.W. Song, S.L. Zhang, L.M. Wu, Detection of DNA using cationic polyhedral oligomeric silsesquioxance nanoparticles as the probe by resonance light scattering technique, Biosens. Bioelectron.22 (2007) 1461-1465. |
| [24] | S. Li, H. Liu, Y. Deng, L. Lin, N. He, Development of a magnetic nanoparticles microarray for simultaneous and simple detection of foodborne pathogens, J. Biomed. Nanotechnol.9 (2013) 1254-1260. |
| [25] | D. Vrbanic, M. Remskar, A. Jesih, et al., Air-stable monodispersed Mo6S3I6 nanowires, Nanotechnology 15 (2004) 635-638. |
| [26] | V. Nicolosi, D. Vrbanic, A. Mrzel, et al., Solubility of Mo6S4.5I4.5 nanowires in common solvents: a sedimentation study, J. Phys. Chem.B 109 (2005) 7124-7133. |
| [27] | V. Nicolosi, D. Vrbanic, A. Mrzel, et al., Solubility of Mo6S4.5I4.5 nanowires, Chem. Phys. Lett.401 (2005) 13-18. |
| [28] | M. Uplaznik, B. Bercic, J. Strle, et al., Conductivity of single Mo6S9-xIx molecular nanowire bundles, Nanotechnology 17 (2006) 5142-5146. |
| [29] | M.I. Ploscaru, S.J. Kokalj, M. Uplaznik, et al., Mo6S9-xIx nanowire recognitive molecular-scale connectivity, Nano Lett. 7 (2007) 1445-1448. |
| [30] | D. Mihailovic, Inorganic molecular wires: physical and functional properties of transition metal chalco-halide polymers, Prog. Mater. Sci. 54 (2009) 309-350. |
| [31] | N.J. Sun, M. McMullan, P. Papakonstantinou, D. Mihailovic, M.X. Li, Amplified optical transduction of proteins derived fromMo6S9-xIx nanowires, Prog. Nat. Sci.: Mater. Int.23 (2013) 326-330. |
| [32] | J. Marmur, A procedure for the isolation of deoxyribonucleic acid from microorganisms, J. Mol. Biol.3 (1961) 208-218. |
| [33] | P. Yang, M.L. Guo, B.S. Yang, Study on the interactions between titanocene dichloride and DNA, Chin. Sci. Bull. 38 (1993) 2049-2052. |
| [34] | Y.M. Song, P.J. Yang, L.F. Wang, M.L. Yang, J.W. Kang, Study on the interactions between Sm(RA)2 Ac 4H2O and DNA, Acta Chim. Sin. 61 (2003) 1266-1270. |
| [35] | E.J. Gao, S.M. Zhao, Q.T. Liu, Study on the interaction of mixed ligand complex palladium(Ⅱ)-biquinoline-phenethylmalonate with DNA, Chin. J. Inorg. Chem. 20 (2004) 191-194. |
| [36] | L.M. Chen, J. Liu, J.C. Chen, et al., Experimental and theoretical studies on the DNAbinding and spectral properties of water-soluble complex Ru(MeIm)4(dpq)]2+, J. Mol. Struct. 881 (2008) 156-166. |
| [37] | Y.Z. Xiang, N. Wang, J. Zhang, et al., Novel cyclen-based linear polymer as a highaffinity binding material forDNA condensation, Sci. China Ser. B 52 (2009) 483-488. |
| [38] | J. Kypr, I. Kejnovská, D. Renčiuk, M. Vorlčková, Circular dichroism and conformational polymorphism of DNA, Nucl. Acids Res.37 (2009) 1713-1725. |
| [39] | R. Chakraborty, S. Chatterjee, S. Sarkar, P. Chattopadhyay, Study of photoinduced interaction between calf thymus-DNA and bovine serum albumin protein with H2Ti3O7 nanotubes, J. Biomater. Nanobiotechnol.3 (2012) 462-468. |
| [40] | S. Wojtulewski, S.J. Grabowski, Different donors and acceptors for intramolecular hydrogen bonds, Chem. Phys. Lett. 378 (2003) 388-394. |
| [41] | A. Soriano, R. Castillo, C. Christov, et al., Catalysis in glycine N-methyltransferase: testing electrostatic stabilization and compression hypothesis, Biochemistry 45 (2006) 14917-14925. |
| [42] | S.B. Novakovic, B. Fraisse, G.A. Bogdanovic, A. Spasojevic-deBire, Experimental charge density evidence for the existence of high polarizability of the electron density of the free electron pairs on the sulfur atom of the thioureido group, NH-C(=S)-NH2, induced by N-H...S and C-H...S interactions, Cryst. Growth Des. 7 (2007) 191-195. |




