b Department of Medical Ultrasonics, The Third Affiliated Hospital, Sun Yat-Sen University, Guangzhou 510630, China;
c MOE Key Laboratory of Laser Life Science & Institute of Laser Life Science, College of Biophotonics, South China Normal University, Guangzhou 510631, China;
d Guangzhou Baiyun District Center for Disease Control & Prevention, Guangzhou 510445, China
Catalytic transfer of genetic information from DNA to RNA is very important in life activities [1],since unconventional RNA transcription levels may be related to the source of genetic diseases. Therefore,the methods for detection of transcriptional levels play a vital role in the interpretation of specific,functional, gene regulated mechanisms,elucidation of the molecular bases of disease caused by gene expression abnormalities,drug target discovery and high throughput drug screening. Reverse transcription polymerase chain reaction (RT-PCR) is the most commonly used method for detection of RNA transcripts. Although RT-PCR is relatively simple and sensitive,electrophoresis detection takes considerable time and requires the use of toxic reagents.
Recently,graphene oxide (GO) has gained tremendous interest in the biosensor field due to its excellent physical and chemical properties,which include a single-layer sheet structure of carbon atoms [2, 3, 4, 5, 6]with carboxylandphenolhydroxylgroupsonitssurface. GO exhibits a different affinity toward single-stranded DNA (ssDNA) versus double-stranded DNA (dsDNA). It can stably adsorb ssDNA whose structure is flexible,and assumes a linear orientation on its surface due to the π–π stacking effect with the nucleic acid bases. WhileintheformationofdsDNA,witharigiddoublehelixstructure,it will have a greatly reduce adsorption ability to the nucleic acid [7, 8, 9]. GO also has a high fluorescence quenching ability. It can be used as a broad-spectrumfluorescencequenching agent.This distinct function is derived from its heterogeneous atomic chemical structure and electronic properties. The sp2 hybrid crystal domain is believed to play leading role in fluorescence quenching [10, 11, 12] and,therefore,a fluorophore,such as SYBRGreen I,can be effectively quenched byGO through fluorescence resonance energy transfer (FRET) mechanism. SYBRGreenI isone of themost sensitive stains available fordetecting dsDNA,which has a faint background fluorescence in the absence of DNA,and has negligible affinity for ssDNA. On the contrary,SYBR Green I exhibits exceptional strong affinity for dsDNA and a large fluorescence enhancement upon dsDNA binding.
In this study,we proposed a GO-based SYBR Green I fluorescence platform for isothermal detection of RNA transcription levels. The platform consists of T7 RNA polymerase transcription mediated amplification of T7 DNA template and GO-based SYBR Green I fluorescent detection of the dsRNA:DNA hybrids composed of the ssRNA transcripts and the label-free ssDNA probes. 2. Experimental 2.1. Materials and reagents
Graphene oxide (1 mg/mL) was purchased from Xianfeng Nanotechnologies Co.,Ltd. (Nanjing,China). Both T7 RNA polymerase and ribonucleotide triphosphate (rNTPs) mix were the products of NewEngland Biolabs (Beijing) Co.,Ltd. SYBR Green I (dsDNA dye) and SYBR Green II (RNA and ssDNA dye) were purchased from Invitrogen Trading (Shanghai) Co.,Ltd. The reagents related to electrophoresis were purchased from Bio-Rad (Richmond,CA). SSC buffer and RNase-freewater were purchased from Shanghai Sangon Biotechnology Co.,Ltd. Oligonucleotides were synthesized and purified by HPLC at Invitrogen Trading (Shanghai) Co.,Ltd. 2.2. T7 RNA transcription system
The transcription system contained T7 DNA template,rNTPs mix (2 mmol/L),T7 RNA polymerase (2.5 U/mL),1× corresponding buffer,and ribonuclease inhibitor (1 U/mL). (The buffer solution is usually stored in the form of high concentration. The concentration of n × stock solution was diluted n times,and form a 1× working solution to use.) The T7 DNA template was obtained by gradient cooling of two ssDNA (denatured at 95 °C for 5 min,then cooling to 25 °C,2 °C per min). Their sequences were 5'-CGCGAAAT-TAATACGACTCACTATAGGGAGA-3 (coding strand) and 5'-GGTTGGTGTGGTTGG-AAAAAAAAAA-TCTCCCTATAGTGAGTCGTATTA- ATTTCGCG-3' (template strand). The transcription mixture was incubated for 2 h at 37 °C. 2.3. Polyacrylamide gel electrophoresis
Transcription products were analyzed on a Bio-Rad slab electrophoresis system (Bio-Rad Laboratories,USA). The 10 mL samples were loaded onto a 12% native polyacrylamide gel (29:1, acryl:bisacryl) in 0.5× tris-borate-EDTA (TBE). Gels were run at room temperature for 1 h at 120 V. The gel was confirmed by SYBR Green I and SYBR Green II staining and photographed by Bio-Rad digital imaging system. 2.4. GO-based SYBR Green I fluorescence detection
The steps were as follows: the ssRNA transcription product was dissolved in 1× SSC buffer,then the label-free ssDNA probe (sequence was 5'-GGTTGGTGTGGTTGG-AAAAAAAAAA-TCTCCC-3') was added to make a final concentration of 50 nmol/L. The mixture was hybridized by incubation for 30 min at 37 °C to produce dsRNA:DNA hybrids. SYBR Green I and GO were then added to make a final concentration of 1× and 8 mg/mL,respectively. After incubating for 10 min,the fluorescence of SYBR Green I binding with the dsRNA:DNA hybrids (excitation at 497 nm and emission at 520 nm) was measured by Perkin-Elmer LS55 luminescence spectrometer (USA). 3. Results and discussion 3.1. Design of the GO-based SYBR Green I fluorescence platform for isothermal detection of RNA transcript levels
This study aims to develop a fast and sensitive tool for isothermal detection of RNA transcript levels. Our proposed method is based on T7 RNA polymerase transcription,nucleic acid hybridization and FRET principle. The T7 RNA polymerase, encoded by T7 bacteriophage [13, 14],is highly specific in its promoter sequence (5'-TAATACGACTCACTATAGGGAGA-3') and is widely used in the isothermal amplification of nucleic acids. In T7 RNA polymerase transcription,the dsDNA template is necessary for the promoter region,but the transcription region only needs ssDNA as the template,therefore in this study,ssDNA template is employed in the transcribed region. The experimental principle is shown in Fig. 1. Briefly,T7 RNA polymerase binds to the promoter region of the T7 DNA template,to begin the transcription reaction, thus producing many copies of ssRNA transcripts,which can hybridize with a label-free ssDNA probe. The probe alone is mainly in the unfolded and flexible state. GO exhibits a different affinity toward ssDNA versus dsDNA and it also has a high fluorescence quenching ability. When the probe is incubated with SYBR Green I and GO, the probe will be adsorbed on the GO surface and the faint background fluorescence of SYBR Green I is largely quenched by GO through the FRET mechanism. The hybridization of the ssRNA transcripts with the probe will produce rigid and definite tertiary structural ds-RNA:DNA hybrids. Similar to the dsDNA, the affinity of this rigid dsRNA:DNA hybrid structure toward GO is very weak, and thus the dsRNA:DNA hybrid will be released from the GO surface. The binding of SYBR Green I with the released dsRNA:DNA hybrid will lead to a large fluorescence enhancement. Therefore, once the dsRNA:DNA hybrid is produced, the hybrid binding with SYBR Green I will emit enhanced fluorescence for detection.

|
Download:
|
| Fig. 1.The principle of monitoring of RNA transcription levels using GO-based SYBR Green I fluorescence platform. | |
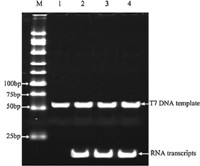
In order to verify the validity of the T7 transcription reaction, 50 nmol/L of T7 DNA template was transcribed by T7 RNA polymerase. The transcripts were detected using 12% polyacrylamide gel electrophoresis. Fig. 2 shows that the RNA transcripts can be stably produced from three parallel transcription experiments.
|
Download:
|
| Fig. 2.Electrophoresis identification of the T7 transcription reaction. M: DNA marker; 1: T7 DNA template; 2–4: three parallel transcription experiments. | |
In order to verify the sensitivity of the GO-based SYBR Green I fluorescence platform,different concentrations of T7 DNA template ranging from 0.5 pmol/L to 50 nmol/L were detected by the assay. The results are shown in Fig. 3A. When the T7 DNA template concentration is 0.5 pmol/L,similar fluorescence intensities are observed between the experimental group and control group. However,with 5 pmol/L of the T7 DNA template,the fluorescence intensity was significantly enhanced,therefore, demonstrating that the currently developed platform achieved a sensitivity of 5 pmol/L.

|
Download:
|
| Fig. 3.Fluorescence spectroscopy experiments for measuring transcripts levels derived from different concentrations of T7 DNA template. (A) Real-time fluorescence spectroscopy of transcripts derived from different concentrations of T7 DNA template. (B) Calibration curve for T7 DNA template. Each point represents the average of fluorescence intensities from three independent assays. Error bar represents the standard deviation from three independent assays. | |
To prepare the calibration standards,three separate samples for each concentration of the T7 DNA template (0.5 pmol/L to 50 nmol/ L) were prepared and analyzed by the platform,respectively,and the averages and standard deviations were calculated using Microsoft Excel spread sheet function. The calibration curve was plotted as the fluorescence intensity against the template concentrations. The calibration curve for T7 DNA template is shown in Fig. 3B. Each point represents the average of fluorescence intensities from three independent assays and the error bar represents the standard deviation from three independent assays. Fig. 3B shows that the fluorescence intensities increase with the template concentrations. The dynamic range of the fluorescence intensity covers the template concentrations from 0.5 pmol/L to 5 nmol/L. However,when the template concentration is greater than 5 nmol/L,a prozone effect occurs and the fluorescence intensity does not rise linearly with the increased template concentration,suggesting that 5 nmol/L template has reached the saturation limit of the GO-based SYBR Green I fluorescence platform.
A good stability and reproducibility of the proposed method can also be observed from the standard deviation from three independent assays in Fig. 3B. 4. Conclusion
In this study,we developed a new GO-based SYBR Green I fluorescence platform for the isothermal detection of RNA transcription levels. The method can clearly discriminate 5 pmol/L of T7 DNA template from the negative control. Acalibration curve with a linearity range from 0.5 pmol/L to 5 nmol/L is established,thus,make quantitative analysis possible. The method may become a powerful tool for RNA transcription detection due to its sensitivity,rapidity and convenience,and may be used for the early detection of the diseases caused by gene expression abnormality. Acknowledgments
This research is supported by the National Natural Science Foundation of China (Nos. 81071790,81371877,61177077),the Key Project of Chinese Ministry of Education (No. 211131),the Science and Technology Project of Guangzhou Nansha District (No. RG201001003),and the Postdoctoral Foundation of China (No. 201003359).
| [1] | W.J. Blake, M. Kaern, C.R. Cantor, J.J. Collins, Noise in eukaryotic gene expression, Nature 422 (2003) 633-637. |
| [2] | K. Lü, G.X. Zhao, X.K. Wang, A brief review of graphene-based material synthesis and its application in environmental pollution management, Chin. Sci. Bull. 57 (2012) 1223-1234. |
| [3] | Z.B. Liu, X.L. Zhang, X.Q. Yan, Y.S. Chen, J.G. Tian, Nonlinear optical properties of graphene-based materials, Chin. Sci. Bull. 57 (2012) 2971-2982. |
| [4] | M.S. Xu, Y. Gao, X. Yang, H.Z. Chen, Unique synthesis of graphene-based materials for clean energy and biological sensing applications, Chin. Sci. Bull. 57 (2012) 3000-3009. |
| [5] | H.Y. Hea, J. Klinowskia, M. Forsterb, A. Lerf, A new structural model for graphite oxide, Chem. Phys. Lett. 287 (1998) 53-56. |
| [6] | P.S. Lau, B.K. Coombes, Y.F. Li, A general approach to the construction of structureswitching reporters from RNA aptamers, Angew. Chem. Int. Ed. 49 (2010) 7938-7942. |
| [7] | C.H. Lu, H.H. Yang, C.L. Zhu, X. Chen, G.N. Chen, A graphene platform for sensing biomolecules, Angew. Chem. Int. Ed. 48 (2009) 4785-4787. |
| [8] | S.J. He, B. Song, D. Li, et al., A graphene nanoprobe for rapid, sensitive, and multicolor fluorescent DNA analysis, Adv. Funct. Mater. 20 (2010) 453-459. |
| [9] | F. Li, Y. Huang, Q. Yang, et al., A graphene-enhanced molecular beacon for homogeneous DNA detection, Nanoscale 2 (2010) 1021-1026. |
| [10] | Y. Wang, Z.H. Li, D.H. Hu, et al., Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells, J. Am. Chem. Soc. 132 (2010) 9274-9276. |
| [11] | H.F. Dong, W.C. Gao, F. Yan, et al., Fluorescence resonance energy transfer between quantum dots and graphene oxide for sensing biomolecules, Anal. Chem. 82 (2010) 5511-5517. |
| [12] | Y.Q. Wen, F.F. Xing, S.J. He, et al., A graphene-based fluorescent nanoprobe for silver (I) ions detection by using graphene oxide and a silver specific oligonucleotide, Chem. Commun. 46 (2010) 2596-2598. |
| [13] | S. Tabor, C. Richardson, A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes, Proc. Natl. Acad. Sci. U. S. A. 82 (1985) 1074-1078. |
| [14] | J.H. Kim, R.G. Larson, Single-molecule analysis of 1D diffusion and transcription elongation of T7 RNA polymerase along individual stretched DNA molecules, Nucleic Acids Res. 35 (2007) 3848-3858. |




