2. 上海海洋大学 港航生态安全研究中心, 上海 201306
相较于多细胞动物,微生物对于生存环境更具适应性,能够普遍存在于生物圈,甚至于某些极端环境[1]。海水中微生物的数量远远超过桡足类、鱼类等大型水生生物,细菌和病毒的数量级达到106~1011个/L[2-5]。据统计,通过船舶压载水途径进行传播的细菌高达1 019 cells/d,数量远远超过其他任何物种[6]。船舶达到目的港口后,微生物一旦被释放到新的环境,将很容易成为入侵物种。
1992年首次在船舶压载水中发现致病型霍乱弧菌,从而揭示了船舶压载水途径传播致病菌的危害性[7]。为防止病原微生物在我国沿海港口传播和扩散,对到港船舶压载水中细菌总数、大肠菌群指标、沙门氏菌指标等卫生管理指示菌以及弧菌等致病菌进行了目的性检测[8-10]。然而,由于缺乏对压载水中微生物种类组成的研究,很难进一步了解压载水中微生物群落结构。高通量测序技术的发展突破了技术手段上的局限性,国内外研究人员得以从更为精准的分类水平进行分析,深入开展到港船舶压载水中微生物多样性和群落结构特征的研究调查[11-14]。但目前相关资料依然较少,对于压载过程中压载舱内微生物种类组成和群落结构变化特征的研究仍是空白。本文应用岸基模拟压载舱,连续跟踪压载后细菌的动态变化情况,以期阐明压载水中细菌的多样性和群落结构的变化规律及其生态适应机制。
1 材料与方法 1.1 样品采集上海海洋大学船舶压载水检测实验室岸基试验基地配备600m3全封闭钢筋混凝土结构模拟压载舱。舱体内壁涂有船舱专用防腐漆,全舱不透光且具有较好隔热性,仅在舱体上部设有直径为100 cm的闭合式人孔,供人员进出和采样所用。设置自然条件下的开放式舱体为对照舱,舱体为600 m3全开放式钢筋混凝土结构。实验前对两舱进行彻底清洗。通过潜水泵引洋山港海域海水,经加载操作注满舱体。完成加载操作后,分别于第0,1,5,10,15和30天进行样品T0,T1,T5,T10,T15,T30的采集,第0天为压载当天,记为样品T0,作为实验起始。同时进行对照舱样本C0,C1,C5,C10,C15和C30的采集。
1.2 样品预处理所有样本均保存于冷藏箱中,2 h内送到实验室, 立即进行预处理。水样通过3 μm和0.22 μm微孔滤膜两级过滤后,将0.22 μm滤膜转移到1.5 mL离心管中,并保存于-80 ℃条件下,用于后续DNA提取。
1.3 基因组DNA提取DNA的提取采用3S DNA Isolation Kit V2.2(上海博彩生物技术有限公司)DNA提取试剂盒,按照操作说明书进行,用1%的琼脂糖凝胶电泳(TAE缓冲液)检测DNA完整性,超微量分光光度计(Nanodrop 2000c,Thermo)分析DNA相对浓度。
1.4 PCR扩增及高通量测序利用16S rRNA基因中V4-V5区域通用引物515F(5’- GTGCCAGCMGCCGCGG-3’)和907R(5’-CCGTCAATTCMTTTRAGTTT-3’)进行PCR扩增。扩增条件设置为:95℃ 3 min,95 ℃ 30 s,5 ℃ 30 s,72 ℃ 5 s,30个循环;72℃延伸10 min。2%琼脂糖凝胶电泳检测PCR产物。PCR产物于上海美吉生物医药科技有限公司Illumina MiSeq平台上进行高通量测序。
1.5 数据分析运用QIIME 1.7.0对Illumina平台测序得到的序列数据进行过滤[15],质控校准后得到优质序列用于后续分析。序列经过滤后,共获得413 806条高质量序列,各样品抽平处理至26 538条序列。在97%的相似水平下对所有序列进行OTU(Operational Taxonomic Units)划分,然后采用RDP classifier贝叶斯算法对97%相似水平的OTU代表序列进行分类学分析,数据库为Silva 11. 9[16]。应用Mothur软件计算生物多样性指数,包括:群落丰富指数(Chao指数)和群落多样性指数(Shannon指数)[17]。运用R语言工具统计和作图软件进行主坐标分析(Principal co-ordinates analysis, PCoA)和非度量多维尺度分析(Non-metric Multidimensional scaling, NMDS)[18]。采用非加权组平均法(Unweighted pair group method with arithmetic mean, UPGMA)构建树状结构。
2 结果与分析 2.1 测序结果和多样性指数通过对实验舱和对照舱各样本点细菌Miseq测序,过滤掉低质量序列后,共获得有效序列413 806条。其中实验舱各样本点平均有效序列数为32 963 ± 4 745,对照舱各样本点平均有效序列数为36 005 ± 3 587。从稀释曲线(图 1)中可知,当各样本序列数量达到30 000后曲线趋于平缓,说明测序量合理,能够真实反映水体中细菌群落情况。Chao和Shannon指数在实验舱样本中的平均值为591.67和3.64,而在对照舱样本中分别为408.00和3.04,说明压载环境下细菌多样性和丰度均有所提高(表 1)。
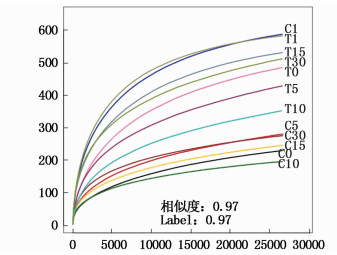
|
图 1 相似度为0.97条件下各样品的稀释曲线
Fig. 1 Rarefaction curves of each sample at cutoff level of 3%
|
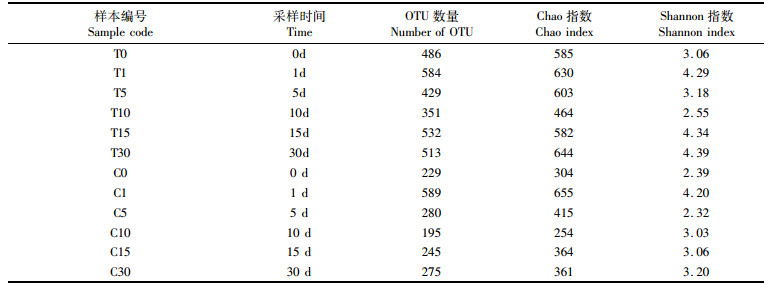
|
表 1 样品测序数据统计表 Tab.1 Sequencing data statistics of samples |
在0.97相似度下得到样本OTU数目,通过维恩图表现实验舱和对照舱细菌群落OTU数目组成相似性、差异性和重叠情况(图 2)。压载环境与自然环境下水体之间存在共有细菌OTU数目为623,实验舱水样中特异性细菌OTU数目为475,是对照舱水样中特异性细菌OTU数目的2.39倍。表明压载条件下有更多的特异性细菌类群存在。

|
图 2 细菌OTU Venn图
Fig. 2 Venn diagram of bacterial OTUs
|
变形菌门(Proteobacteria)为两舱体内主要细菌类群,实验舱中所占比例为66.23%±13.31%,对照舱为53.17%±10.99%(图 3)。在发现的变形菌门中包括α-、β-、γ-、δ-变形菌,其中γ-变形菌纲占据菌群主要优势。实验舱中γ-变形菌的丰度比例范围为27.29%~73.43%,由第0天时40.60%缓慢下降至第5天时32.33%,在第10天迅速增加至73.43%;此时,对照舱内γ-变形菌比例仅为8.18%,而α-变形菌纲丰度比例达到24.11%(第5天时仅6.36%)。拟杆菌门(Bacteroidetes)丰度比例在实验舱中的变化范围为9.45%~45.76%,在对照舱为19.38%~66.48%,是出现的第二大类群细菌。其次是放线菌门(Actinobacteria),丰度比例在实验舱的变化范围为1.68%~10.76%,对照舱是0.37%~27.18%。变形菌门和拟杆菌门细菌在两舱中均表现出绝对的类群优势。酸杆菌门(Acidobacteria)和绿弯菌门(Chloroflexi)是实验舱压载阶段出现的特有门类,压载前期(10 d前)丰度比例低于1%,压载第30天时两者丰度增加至2.19%和3.69%(图 3)。蓝细菌门(Cyanobacteria)则仅在对照舱中出现。
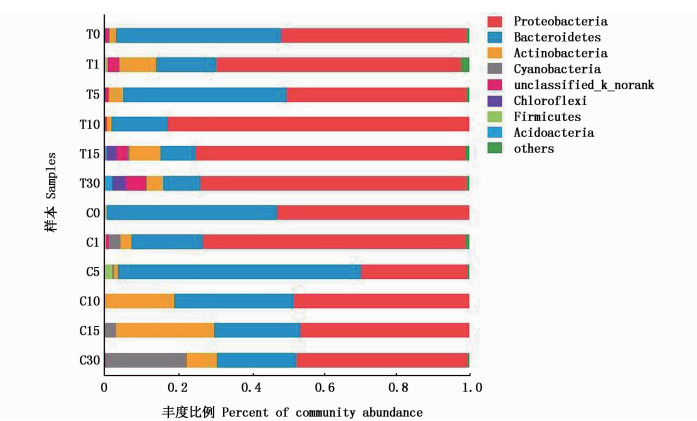
|
图 3 细菌群落在门水平上的组成和相对丰度
Fig. 3 Composition at the phylum level and relative abundance of bacterial communities
|
在科的分类水平上,黄杆菌科(Flavobacteriaceae)是实验初始的优势分类单元,第0天时实验舱和对照舱中的相对比例分别是43.52%和46.33% (图 4)。黄杆菌科(Flavobacteriaceae)在两舱内的相对丰度比例均表现出先增加后减少的变化趋势,第5天时上升至最高比例后逐渐下降。假替单胞菌科(Pseudoalteromonadaceae)在实验舱内的比例在第10天急剧增加至50.16%,随后菌群优势减弱。对照舱中微杆菌科(Microbacteriaceae)、Methylophilaceae和Surface_1的丰度比例增长尤为显著。
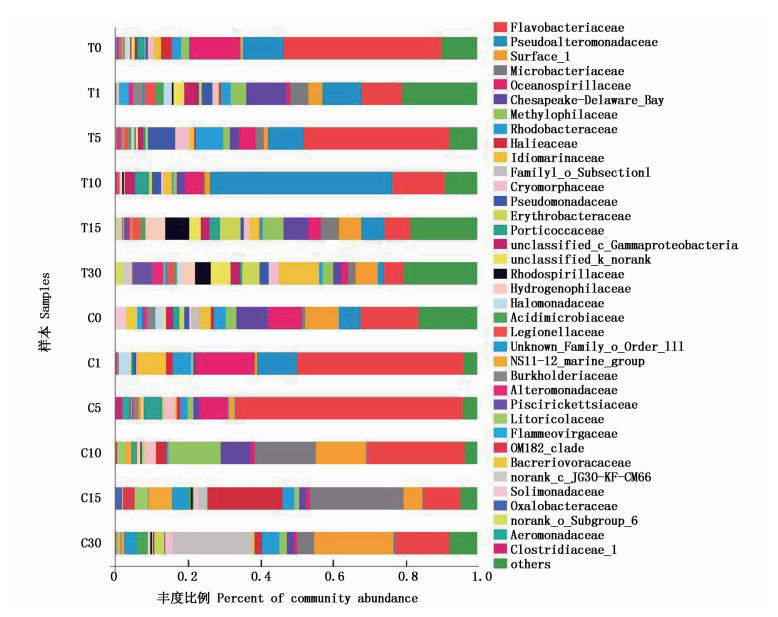
|
图 4 细菌群落在科水平上的组成和相对丰度
Fig. 4 Composition at the family level and relative abundance of bacterial communities
|
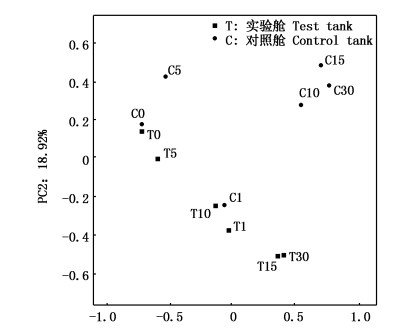
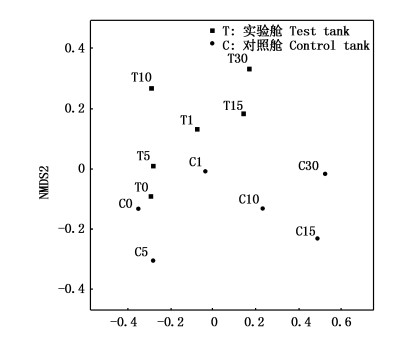
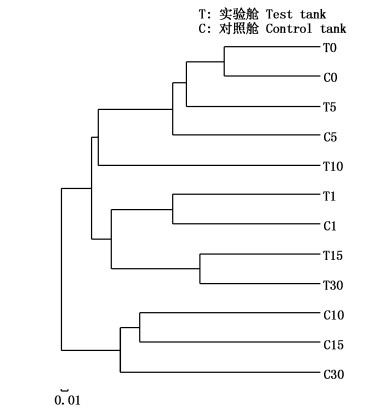
PCoA分析用于研究样本群落组成的相似性或差异性,直观显示实验舱和对照舱细菌群落变化上的相似性和差异性(图 5)。PC1和PC2两轴分别解释了18.92%和42.87%的细菌群落变化,第0天、1天两舱体细菌群落接近,此后相对距离增大,表明不同环境下细菌群落变化情况的差异性。NMDS分析同样揭示了两舱体内细菌群落结构的不同(图 6)。基于层次聚类中的非加权组平均法UPGMA对各采样点样品细菌群落构成的相似性进行了聚类分析,对照舱各采样点相似度较高,实验舱内细菌在压载过程中的群落结构表现出较大的波动,因此受到压载环境的显著影响(图 7)。
|
图 5 细菌群落PCoA分析
Fig. 5 Principal co-ordinates analysis between samples
|
|
图 6 细菌群落NMDS分析
Fig. 6 Non-metric multidimensional scaling analysis between samples
|
|
图 7 多样本相似度树状图
Fig. 7 Hierarchical clustering by unweighted pair group method with arithmetic mean (UPGMA) according to their weighted unifrac matrix.
|
通过对压载水中细菌群落进行连续压载30 d的Miseq高通量测序,结果表明细菌群落的丰度和多样性在一定程度上随压载时间的延长而增加(表 1)。一般在稳定良好的生态环境中,微生物数量增加,种群丰富度上升,意味着群落结构表现更为稳定[19]。在压载舱内,温度突变、黑暗密闭、无外来能源供给、舱壁防污涂料的毒性侵害等恶劣的压载舱条件使得大多数浮游植物和浮游动物无法长时间存活,经过长时间压载后种类数量以及丰度将会明显下降[20-21]。然而,压载水中细菌的丰度及其生物量能够保持稳定,与加载地水源保持一致,甚至发生随压载时间不断增长的变化情况[22-23]。HUA等连续跟踪4条航线商船压载水中可培细菌总数的变化情况,在他所调查的6艘船舶航行过程中,细菌总数均有增加的情况,尤其发生在较长水龄时期[24]。TOMARU等发现,压载水交换操作后可培细菌总数减少,但在短时下降之后,数量又随压载时间的延长而不断上升[25]。尽管以上研究中实验方法与本次研究不同,在具体数量上无法作出比较,但都在一定程度上证实了压载水中的微生物具有较高的存活能力,相较于其他浮游生物更易对目的港口造成生物入侵。
本次岸基实验压载水源来自洋山港近岸海域,该水域细菌主要由变形菌、拟杆菌这两大类组成,其中γ-变形菌为优势类群[26]。相较于对照舱,γ-变形菌在实验舱内的丰度优势更为明显,压载过程中比例变化以升高为主。蓝细菌依靠光合作用产生能量维持生长,光照是影响其生长的重要因素[27],压载舱黑暗无光的环境抑制了蓝细菌的生长,导致在压载水样品中蓝藻的丰度极低。变形菌门细菌呈革兰氏阴性,外膜结构主要由脂多糖组成。该类细菌包含多种代谢途径,以营兼性或者专性厌氧及异养为主要生活方式。黄杆菌科(Flavobacteriia)和假替单胞菌科(Pseudoalteromonadaceae)是实验舱中γ-变形菌优势类群,其中黄杆菌科为好氧或兼性好氧的化能异养型微生物[28],对环境适应能力较强;假替单胞菌科对于蛋白质和脂肪的分解能力极强,且增殖速度快,浮游生物大量死亡后,该类群细菌丰度迅速增加(图 4)。
此外,在实验舱样本中存在一定比例的无法按目前分类学研究划分的菌属,说明在压载环境中有一定数量潜在的新种和细菌分类(图 3)。并且此类群菌属的丰度比例随压载时间不断增大,由0.51%(5 d)上升至5.43%(30 d)。压载舱是区别于其他任何自然条件的特殊环境,该环境对于微生物的生理生化特征以及代谢机理都可能造成影响。XU等曾在浙江舟山到港船舶压载舱沉积物中成功分离鉴定新菌Kordiimnas lacus[29]。压载水较沉积物而言,有更高的细菌丰度和多样性[30],因此具有更为丰富开发潜力和更高的入侵风险。若船舶压载水未经过处理直接排放,极易造成港口水域生态环境的污染,同时引发病原微生物污染,对人类健康造成威胁。
压载水因其携带致病微生物排放至港口国而获得重视。在以往的研究中,弧菌、绿脓杆菌和分支杆菌等致病菌或条件致病菌均有报道[31-33],其中分支杆菌在实验舱各采样点均有发现。ROBIN在2016年通过高通量测序技术在压载水中发现了60种新的致病菌和条件致病菌,其中包括47种针对人体产生危害的病原菌[34]。泡囊短波单胞菌(Brevundimonas vesicularis)是一种较罕见的条件致病菌,可引发肺炎、败血症、皮肤炎症等病症[35]。本次岸基模拟压载实验中,泡囊短波单胞菌在实验舱内的丰度比例随压载时间持续增加,30 d时达到1.14%。假单胞菌属(Pseudomonas sp.)是常见的病菌属,在实验舱中共检出6种(包括1种无法确定种类)。某些功能细菌可能产生某些活性物质,例如Pseudomonas xanthomarina是仅有的能够对有机化合物进行降解的细菌[35],在实验舱内的丰度最高达7.02%,这意味着压载环境下蕴藏很大的开发潜力。但与此同时,压载环境下也可能存在新型病源微生物,这对于人类健康以及其他生物的生存都存在巨大的风险。
| [1] |
DEMING J W. Unusual or extreme high-pressure marine environments[M]//HURST C J, KNUDSEN G R, MCINERNEY M J, et al. Manual of Environmental Microbiology. Washington, DC: ASM Press, 1997: 478-490.
|
| [2] |
DUCKLOW H W, SHIAH F K. Estuarine bacterial production[M]//FORD T E. Aquatic Microbiology: An Ecological Approach. America: Blackwell, 1993: 261-284.
|
| [3] |
PROCTOR L M. Advances in the study of marine viruses[J]. Microscopy Research&Technique, 1997, 37(2): 136-161. |
| [4] |
FUHRMAN J A. Marine viruses and their biogeochemical and ecological effects[J]. Nature, 1999(6736): 541-548. |
| [5] |
WOMMACK K E, COLWELL R R. Virioplankton:viruses in aquatic ecosystems[J]. Microbiology and Molecular Biology Reviews, 2000, 64(1): 69-114. DOI:10.1128/MMBR.64.1.69-114.2000 |
| [6] |
RUIZ G M, RAWLINGS T K, DOBBS F C, et al. Global spread of microorganisms by ships[J]. Nature, 2000, 408(6808): 49-50. DOI:10.1038/35040695 |
| [7] |
MCCARTHY S A, MCPHEARSON R M, GUARINO A M, et al. Toxigenic Vibrio cholerae 01 and cargo ships entering Gulf of Mexico[J]. The Lancet, 1992, 339(8793): 624-625. |
| [8] |
赵丽青, 贾俊涛, 姜英辉, 等. 山东主要港口海域及入境船舶压舱水中病原菌调查结果的多维尺度分析[J]. 中国国境卫生检疫杂志, 2013, 36(5): 334-337. ZHAO L Q, JIA J T, JIANG Y H, et al. MDS analysis on the pathogenic bacterias in Shandong main Harbor water and entry ship ballast water[J]. Chinese Journal of Frontier Health and Quarantine, 2013, 36(5): 334-337. |
| [9] |
李春丽, 刘兵, 周君, 等. 宁波港压载水的细菌多样性研究[J]. 生态科学, 2012, 31(6): 636-644, 670. LI C L, LIU B, ZHOU J, et al. Study on genetic diversity of bacteria in ballast water from Ningbo port[J]. Ecological Science, 2012, 31(6): 636-644, 670. |
| [10] |
冯云霄, 张乐, 方振东, 等. 秦皇岛口岸入境船舶压载水中微生物携带情况调查[J]. 环境科学导刊, 2011, 30(2): 58-61. FENG Y X, ZHANG L, FANG Z D, et al. Investigation of microbe taken by ballast water of entry ships in Qinhuangdao port[J]. Environmental Science Survey, 2011, 30(2): 58-61. |
| [11] |
FUJIMOTO M, MOYERBRAILEAN G A, NOMAN S, et al. Application of ion torrent sequencing to the assessment of the effect of alkali ballast water treatment on microbial community diversity[J]. PLoS ONE, 2014, 9(9): e107534. DOI:10.1371/journal.pone.0107534 |
| [12] |
NG C, LE T H, GOH S G, et al. A comparison of microbial water quality and diversity for ballast and tropical harbour waters[J]. PLoS ONE, 2015, 10(11): e0143123. DOI:10.1371/journal.pone.0143123 |
| [13] |
BRINKMEYER R. Diversity of bacteria in ships ballast water as revealed by next generation DNA sequencing[J]. Marine Pollution Bulletin, 2016, 107(1): 277-285. DOI:10.1016/j.marpolbul.2016.03.058 |
| [14] |
WU H, CHEN C, WANG Q, et al. The biological content of ballast water in China:A review[J]. Aquaculture & Fisheries, 2017, 2(6): 241-246. |
| [15] |
CAPORASO J G, KUCZYNSKI J, STOMBAUGH J, et al. QⅡME allows analysis of high-throughput community sequencing data[J]. Nature Methods, 2010, 7(5): 335-336. DOI:10.1038/nmeth.f.303 |
| [16] |
QUAST C, PRUESSE E, YILMAZ P, et al. The SILÜA ribosomal RNA gene database project:improved data processing and web-based tools[J]. Nucleic Acids Research, 2013, 41(Database issue): D590-D596. |
| [17] |
SCHLOSS P D, GEVERS D, WESTCOTT S L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies[J]. PLoS ONE, 2001, 6(12): e27310. |
| [18] |
RIVAS M N, BURTON O T, WISE P, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis[J]. Journal of Allergy and Clinical Immunology, 2013, 131(1): 201-212. DOI:10.1016/j.jaci.2012.10.026 |
| [19] |
贺纪正, 李晶, 郑袁明. 土壤生态系统微生物多样性-稳定性关系的思考[J]. 生物多样性, 2013, 21(4): 411-420. HE J Z, LI J, ZHENG Y M. Thoughts on the microbial diversity-stability relationship in soil ecosystems[J]. Biodiversity Science, 2013, 21(4): 411-420. |
| [20] |
GOLLASCH S, LENZ J, DAMMER M, et al. Survival of tropical ballast water organisms during a cruise from the Indian Ocean to the North Sea[J]. Journal of Plankton Research, 2000, 22(5): 923-937. DOI:10.1093/plankt/22.5.923 |
| [21] |
CHAN F T, BRISKI E, BAILEY S A, et al. Richness-abundance relationships for zooplankton in ballast water:temperate versus Arctic Comparisons[J]. ICES Journal of Marine Science, 2014, 71(7): 1876-1884. DOI:10.1093/icesjms/fsu020 |
| [22] |
SEIDEN J M, WAY C, RIVKIN R B. Microbial hitchhikers:Dynamics of bacterial populations in ballast water during a trans-Pacific voyage of a bulk carrier[J]. Aquatic Invasions, 2010, 5(1): 13-22. DOI:10.3391/ai |
| [23] |
HUA J, HWANG W H. Effects of voyage routing on the survival of microbes in ballast water[J]. Ocean Engineering, 2012, 42: 165-175. DOI:10.1016/j.oceaneng.2012.01.013 |
| [24] |
TOMARU A, KAWACHI M, DEMURA M, et al. Changes in microbial communities, including both uncultured and culturable bacteria, with mid-ocean ballast-water exchange during a voyage from Japan to Australia[J]. PLoS ONE, 2014, 9(5): e96274. DOI:10.1371/journal.pone.0096274 |
| [25] |
薛俊增, 肖南燕, 王琼, 等. 洋山港海域细菌群落多样性的季节变化[J]. 生态学报, 2016, 36(23): 7758-7767. XUE J Z, XIAO N Y, WANG Q, et al. Seasonal variation of bacterial community diversity in Yangshan Port[J]. Acta Ecologica Sinica, 2016, 36(23): 7758-7767. |
| [26] |
HERLEMANN D P, LABRENZ M, JVRGENS K, et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea[J]. Isme Journal, 2011, 5(10): 1571-1579. DOI:10.1038/ismej.2011.41 |
| [27] |
BOONE D R, CASTENHOLZ R W, GARRITY G M, et al. Bergey's Manual of Systemativ Bacteridogy Volume One:the Archaea and the deeply branching and phototrophic bacteria[M]. 2nd ed. New York: Springer Verlag, 2001.
|
| [28] |
XU X W, HUO Y Y, BAI X D, et al. Kordiimonas lacus sp.nov., isolated from a ballast water tank, and emended description of the genus Kordiimonas[J]. International Journal of Systematic and Evolutionary Microbiology, 2011, 61(2): 422-426. DOI:10.1099/ijs.0.018200-0 |
| [29] |
DRAKE L A, DOBLIN M A, DOBBS F C. Potential microbial bioinvasions via ships' ballast water, sediment, and biofilm[J]. Marine Pollution Bulletin, 2007, 55(7-9): 333-341. DOI:10.1016/j.marpolbul.2006.11.007 |
| [30] |
ALTUG G, GURUN S, CARDAK M, et al. The occurrence of pathogenic bacteria in some ships' ballast water incoming from various marine regions to the Sea of Marmara, Turkey[J]. Marine Environmental Research, 2012, 81: 35-42. DOI:10.1016/j.marenvres.2012.08.005 |
| [31] |
BURKHOLDER J M, HALLEGRAEFF G M, MELIA G, et al. Phytoplankton and bacterial assemblages in ballast water of U.S. military ships as a function of port of origin, voyage time, and ocean exchange practices[J]. Harmful Algae, 2007, 6(4): 486-518. DOI:10.1016/j.hal.2006.11.006 |
| [32] |
EMAMI K, ASKARI V, ULLRICH M, et al. Characterization of bacteria in ballast water using MALDI-TOF mass spectrometry[J]. Plos One, 2012, 7(6): e38515. DOI:10.1371/journal.pone.0038515 |
| [33] |
BRINKMEYER R. Diversity of bacteria in ships ballast water as revealed by next generation DNA sequencing[J]. Marine Pollution Bulletin, 2016, 107(1): 277-285. DOI:10.1016/j.marpolbul.2016.03.058 |
| [34] |
CHANDRA A B, CHANDRA P A, CHAPNICK E K. Bacteremia caused by Brevundimonas vesicularis in a patient with biliary pancreatitis[J]. Infectious Diseases in Clinical Practice, 2010, 18(1): 54-55. DOI:10.1097/IPC.0b013e3181a4c87e |
| [35] |
SOPEÑA F, LAIZ L, MORILLO E, et al. Phenanthrene biodegradation by Pseudomonas xanthomarina isolated from an aged contaminated soil[J]. Clean-soil Air Water, 2014, 42(6): 785-790. DOI:10.1002/clen.v42.6 |
2. Centre for Research on the Ecological Security of Ports and Shipping, Shanghai 201306, China
 2018,
Vol. 27
2018,
Vol. 27


