扩展功能
文章信息
- 牛黛醇, 李效宇
- NIU Daichun, LI Xiaoyu
- MicroRNA在鱼类胚胎发育中的调控作用
- The Roles of MicroRNA in Fish Embryonic Development
- 四川动物, 2015, 34(6): 948-954
- Sichuan Journal of Zoology, 2015, 34(6): 948-954
- 10.11984/j.issn.1000-7083.20150091
-
文章历史
- 收稿日期: 2015-03-12
- 接受日期: 2015-05-08
MicroRNA(miRNA)是一类长度为21~23个核苷酸的非编码小分子RNA,一般来源于染色体的非编码区域,由大约70个核苷酸大小的、可形成发夹结构的前体加工而来(Ambros,2004;Bartel,2004; He & Hannon,2004;郝克红等,2011)。1993年,Lee等在秀丽隐杆线虫中意外发现了首个miRNA家族成员lin-4,它是一种定时调控胚胎发育后期的、长度约为22 nt的非编码RNA(Lee et al.,1993;Wightman et al.,1993)。随后几年中,关于miRNA分子的研究几乎没有任何进展。直到2000年,Reinhart等在线虫中发现了另一种重要的、具有转录后调节作用的miRNA——let-7,其表达促进晚期幼虫向成虫过渡(Reinhart et al.,2000;Pasquinelli et al.,2000)。此后,人们在miRNA领域开展了大量研究,迄今在动植物及病毒中已有数以万计的miRNA成员被发现。
成熟miRNA的生物发生是一个复杂的过程,首先在细胞核内miRNA基因被RNA聚合酶Ⅱ或Ⅲ转录为pri-microRNA的初级转录产物(Borchert et al.,2006),然后pri-microRNA被RNA聚合酶Ⅲ Drosha剪切产生大小约为70个核苷酸并能形成茎-环结构的miRNA前体(pre-microRNA)(Lee et al.,2003),miRNA前体在转运蛋白Exportin-5的介导下从细胞核转移到细胞质中,并被RNA聚合酶Ⅲ Dicer加工成为大小为22个核苷酸的miRNA双链,最后与RNA诱导的基因沉默复合物(RNA-induced silencing complex,RISC)中的Argonaute蛋白结合形成非对称的RISC,进而调控基因的表达(Pillai et al.,2007;Davis & Hata,2009)。
大量研究证实,miRNA主要通过与靶基因3’端非编码区序列的互补配对程度来调控靶基因表达,促进靶基因mRNA降解或抑制转录后的翻译过程。miRNA调控其靶基因的方式有2种(Lakshmipathy & Hart,2008),一种是当miRNA与靶基因完全互补时,miRNA直接导致靶mRNA降解;另一种是当两者不完全互补时,miRNA与靶mRNA的3’端非翻译区结合,阻遏基因转录后的翻译过程(图 1)。最新研究表明,miRNA也可以结合到靶mRNA的5’末端、miRNA前体及DNA而抑制其翻译( Fang & Rajewsky,2011)。在多细胞动物中,每个miRNA可以结合200个靶基因,而每个靶基因可以被多个miRNA识别(Betel et al.,2008)。
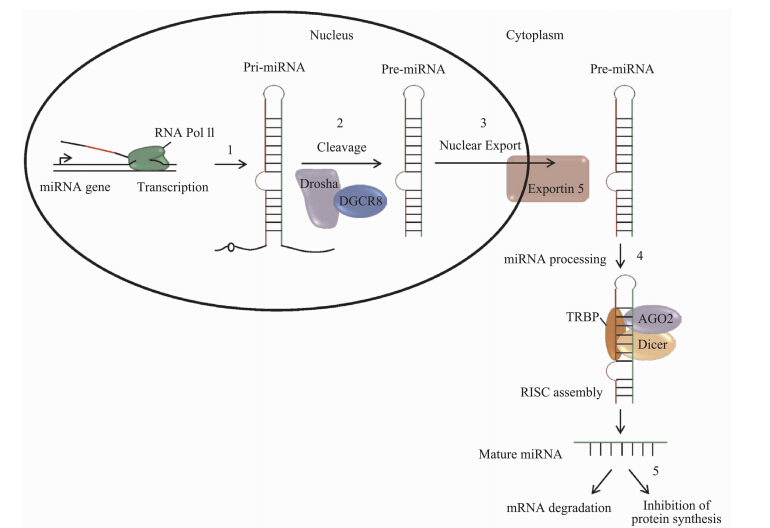 |
| 图 1 miRNA的生物合成及作用机理示意图(Shrivastava et al.,2015) Fig. 1 Sketch of biosynthesis and function mechanism of miRNA(Shrivastava et al.,2015) |
miRNA在进化过程中高度保守,它是生物基因调控网络中的重要成员,可调控约30%的人类基因表达(Lewis et al.,2003)。miRNA参与动植物的生长发育、细胞分化、细胞增殖与凋亡、激素分泌、疾病发生等多种生理和病理过程(Song & Tuan,2006)。miRNA在转录后水平调控组织器官的发育。对鱼类基因组的研究表明,miRNA调控鱼类胚胎发育的全过程。
1 鱼类miRNA鱼类是重要的脊椎动物模式生物之一,现存鱼类超过3万种,约占地球上脊椎动物总物种的一半(IUCN,2011)。近年来,关于脊椎动物中miRNA的研究多有报道,但与其他脊椎动物相比,鱼类miRNA的研究相对较少(唐雪莲等,2013)。鱼类miRNA的研究最早始于斑马鱼Danio rerio(王佳佳等,2007),而后相继在青鳉Oryzias latipes、红鳍东方鲀Fugu rubripes、绿河鲀Tetraodon nigroviridis、白鲢Hypophthalmichthys molitrix等鱼类中展开(Tani et al.,2010;Chi et al.,2011;Ma et al.,2012)。
牛黛醇等:MicroRNA在鱼类胚胎发育中的调控作用 Wienholds等(2005)利用微阵列和原位杂交技术对斑马鱼胚胎进行研究分析,结果发现115个保守miRNAs在胚胎发育过程中呈高度的组织特异性表达,其中大多数miRNA在体节形成期和胚胎发育中晚期表达,而在早期胚胎发育阶段几乎不表达。近年来关于斑马鱼miRNA的研究进展较快,如丁雷等(2011)对斑马鱼胚胎发育中miRNA的表达进行了详细的描述;Li等(2010)在青鳉中发现了254种保守的miRNAs前体,编码593种成熟miRNAs;Johansen等(2011)对大西洋鳕鱼Gadus morhua胚胎发育阶段和成鱼组织中的转录RNA进行深度测序和生物信息学分析,发现了30种miRNAs的表达。
在牙鲆Paralichthys olivaceus的变态发育过程中,其组织、器官的形成及形态的变化都受到miRNA的调控,Fu等(2011)利用微阵列的方法研究发现,miRNA参与调控牙鲆的变态发育。Xie等(2011)也在牙鲆体内发现了4种新miRNAs(miR-20c、miR-23c、miR-130d、miR-181e),并发现16种miRNAs与其变态发育相关。miRNA在北极嘉鱼Salvelinus alpinus胚胎发育中同样发挥着重要的作用,研究者运用高通量测序的方法对其胚胎发育过程进行研究,发现了326种保守的miRNAs和427种新的miRNAs家族的表达; 进一步研究发现,在其胚胎发育阶段有51种保守的miRNAs和6种新的miRNAs差异表达(Kapralova et al.,2014)。
采用同源序列分析结合小RNA测序,在鲤鱼Cyprinus carpio体内鉴定了92种保守的miRNAs和21种鲤鱼特有的miRNAs,并且保守的miRNA具有相对较高的表达水平(Zhu et al.,2012)。有研究者利用生物信息学方法预测了斑点叉尾鮰Ictalurus punctatus表达的16种miRNAs,并用Real Time-PCR验证这些miRNA在其5种不同组织中的表达情况(Xu et al.,2012)。据报道,在斑点叉尾鮰体内鉴定了237种miRNAs和45种新miRNAs,利用Real Time-PCR技术验证了新miRNAs在斑点叉尾鮰肝脏、肌肉、脑、肾脏等10种组织内的表达情况,发现了部分新的miRNAs,如miR-129b、miR-7562、miR-7553在所有组织均有表达,而部分新miRNAs具有高度的组织特异性(Xu et al.,2013)。在其他一些鱼类中也发现了多种miRNA的表达,如大西洋鲑鱼Salmo salas、鲢鱼、鳙鱼Aristichthys nobili、虹鳟Oncorhynchus mykiss等(Salem et al.,2010;Chi et al.,2011;Bekaert et al.,2013)。对鱼类miRNA的研究表明,miRNA的表达及功能对鱼类的发育、生理过程及疾病防御等发挥着重要的调节作用。
2 miRNA在鱼类胚胎发育中的调控作用鱼类作为脊椎动物的主要模式生物,其miRNA的研究主要集中在表达鉴定和寻找新的miRNA以及靶基因的预测,而关于鱼类miRNA功能的研究较少。根据目前的研究报道,miRNA在鱼类的细胞分化、增殖、胚胎形成、生长发育、各种生理功能等生命进程中具有重要的调控作用(Kerr et al.,2011)。近年来,关于miRNA与组织器官形成的报道越来越多,包括对心脏、血管、肌肉、肺、肾脏、胰腺、皮肤、牙齿、内耳、脑和造血系统等发育的调控作用(郝克红等,2011)。虽然鱼类中miRNA功能的具体调控机制尚不清楚,但其在鱼类胚胎发育过程中调控各组织及器官的形成起着至关重要的作用已毋庸置疑。自从鱼类miRNA首次在斑马鱼胚胎发育中被发现,有关miRNA 在鱼类胚胎发育中调控作用的研究报道迅速增多。
2.1 miRNA在鱼类早期胚胎发育中的调控作用许多miRNA有短暂的表达模式,例如在母型-合子型过渡(MZT)和变态中的表达,MZT包括母型mRNA的消除和合子型开始转录2个过程。它有2种调控网络,即母型与合子型。母型mRNA稳定、翻译和定位是早期胚胎发育调控的3个特点(Tadros & Lipshitz,2009)。鱼类中胚胎基因的激活发生在中囊胚期,Bobe等(2000)研究发现虹鳟中囊胚期发生在受精后2 d。很多研究都发现,miR-430在鱼类胚胎发育的囊胚期高表达,这一发现分别在斑马鱼、青鳉、鳕鱼的研究中得到证实,miR-430可促进母源mRNA脱腺苷化和母源mRNA的清除(Giraldez et al.,2006;Tani et al.,2010;Johansen et al.,2011)。另外,miR-34、miR-200a、miR-200b和miR-206也在鱼类胚胎早期发育中表达。miR-206在斑马鱼的母源基因与合子基因中均有表达,在原肠胚期间它主要控制细胞基本运动(Liu et al.,2012),而miR-34是母源型基因,参与斑马鱼早期中枢神经发育(Soni et al.,2013)。
多项研究表明,miRNA在鱼类胚胎早期发育过程中扮演着重要角色。斑马鱼胚胎与幼体缺失合子型的Dicer1导致发育缓慢及幼体存活时间变短(Wienholds et al.,2003)。在斑马鱼胚胎发育中母型与合子型Dicer突变可使原肠胚形成、大脑形成、体节发生和心脏发育出现异常;但把成熟的miR-430注射到Dicer缺失的胚胎内可减少原肠胚和脑室形态发育的缺陷(Giraldez et al.,2005)。
2.2 miRNA在鱼类大脑形成中的调控作用多种miRNA被发现在鱼类大脑的特定区域表达,这表明在一个特定的区域miRNA具有抑制功能。在鱼类中,大脑特异miRNA在不同的类群中具有保守性(Xu et al.,2013)。大脑miRNA的表达依赖细胞的形态,如miR-92b在增殖的神经细胞中广泛表达,miR-124仅在已分化的神经细胞中表达,而miR-9与miR-135c在增殖的细胞和已分化的细胞中均有表达(Kapsimali et al.,2007)。此外,miRNA有大脑组织表达特异性,如miR-9是鱼类和哺乳动物大脑特异表达miRNA,在胚胎神经管晚期选择性表达。功能性研究表明,miR-9可以通过作用于成纤维生长因子Fgf信号途径来调控中后脑边界器官的组织活性(Leucht et al.,2008),而miR-7缺失可能导致中脑缩小,但不影响大脑顶部的端脑(Memczak et al.,2013)。miR-219过表达能引起斑马鱼胚胎大脑发育中前脑细胞凋亡(张满仓等,2008)。有研究发现,miR-125b也与鱼类大脑发育有关,它作用于p53 mRNA的3’UTR从而抑制P53蛋白的表达,斑马鱼胚胎经γ射线和喜树碱处理后miR-125b的表达下调,P53蛋白水平增加,进而导致脑细胞凋亡(Lim et al.,2009)。综上所述,miRNA在鱼类大脑局部的、短暂的、基本的表达说明它们在大脑形态发生和维持不同区域与细胞形态中具有重要功能。
2.3 miRNA在鱼类眼睛形成中的作用鱼类眼睛胚胎发育的起点类似于其他脊椎动物,鱼类视力依赖一个物种的生态位和行为,眼睛组织特异性基因表达指导这种适应。眼睛发育已经在斑马鱼中通过形态学、基因表达和原位标记方法获得很好的了解(Gestri et al.,2012)。Xu等(2009)研究了miRNA在视网膜和眼睛组织中的表达,建立了视网膜、晶状体和角膜的miRNA转录组,发现许多miRNA显示出组织特异性和发育阶段特异性表达模式。
已有的研究结果表明,miR-96、miR-124a、miR-181a、miR-181b、miR-182、miR-183、miR-184和miR-204均在斑马鱼胚胎眼睛发育过程中有特异的表达(Cavodeassi et al.,2005)。与此类似,有研究者发现,一些miRNA在亚洲尖吻鲈Lates calcarifer眼睛发育中特异表达(Xia et al.,2011)。miRNA的空间定位揭示了细胞形态和发育阶段特异性表达模式,例如miR-181b和miR-181a在视网膜细胞中特异表达(Leucht et al.,2008)。miR-183-1和miR-183-2在尖吻鲈眼睛发育中高表达,miR-183在眼睛、鼻子、内耳的感觉细胞中特异表达(Pierce et al.,2008)。Conte等(2010)采用基因敲除的方法研究青鳉的miRNA,发现miR-204靶向meis2调控pax6转录途径;敲除miR-204导致眼睛发育畸形,包括眼杯损伤、眼胚胎缩小、晶状体发育受损、晶状体上皮细胞缺陷、主要纤维细胞错位和裂解以及视神经裂缝关闭。其他的一些miRNA具有维持眼睛细胞内稳态的作用,如let-7在分化阶段维持神经胶质细胞的稳态(Ramach and ran et al.,2010)。另外,miR-30a、miR-184、miR-92b、miR-9和let-7b也参与鱼类眼睛的发育。
2.4 miRNA在鱼类肌肉形成中的调控作用miRNA具有组织表达特异性,骨骼肌和心肌中miRNA特异表达。miRNA在鱼类肌肉的增殖、分化等发育过程中发挥重要的调节作用,其异常表达常与某些肌肉疾病的病理过程有关。鱼类肌肉形成始于胚胎发育期间前体肌原细胞的形成,这些肌原细胞分化成有4个细胞群的肌节,即肌肉前体、慢肌、快肌和内侧肌(Johnston,2006)。这些细胞簇被肌原性调控因子调控,MyoD是一种肌原性调控因子,调控几种下游基因参与肌肉形成,在尼罗河罗非鱼Oreochromis niloticus中MyoD被miR-203b直接调控(Yan et al.,2013)。在鲤鱼骨骼肌中已经发现了几种miRNA的表达,其中一些仅在鱼类中表达。 miR-1、miR-133和miR-206是肌肉特异性miRNA,调控成肌细胞的分化和增殖(Townley-Tilson et al.,2010),其中miR-1和miR-133参与斑马鱼胚胎中肌动蛋白组织的形成(Mishima et al.,2009),miR-1在尖吻鲈肌肉形成中高表达(Pierce et al.,2008)。
在骨骼肌形成中,成肌细胞分化成慢肌细胞纤维和快肌细胞纤维。miR-499通过抑制Sox6的表达导致慢肌细胞纤维形成和维持,而促进快肌细胞纤维分化,这种机制在脊椎动物中是保守的(Wang et al. ,2011)。miR-30表达抑制增加Hedgehog信号的活性,导致斑马鱼慢肌细胞纤维数目增加(Ketley et al.,2013);而抑制斑马鱼肌肉中miR-214表达,增强肌肉细胞对Hedgehog信号的应答,导致慢肌细胞纤维数目减少(Flynt et al.,2007)。在尼罗河罗非鱼骨骼肌发育过程中有4种miRNA(miR-1、miR-27a、miR-133a、miR-206)呈现差异表达(Yan et al.,2012a)。与鱼类肌肉发育相关的miRNA还有let-7、miR-19和miR-130等,而进一步研究单一miRNA的功能对于了解鱼类肌肉形成具有至关重要的作用。
2.5 miRNA在鱼类心血管形成中的调控作用在鱼类器官形成的开始阶段,心脏搏动是一个可辨识的发育阶段,因为它的可见性优于其他的一些可辨识器官。所有后续的生命事件都依赖心脏连续的收缩性。心脏发育是一个非常复杂的过程,区分2种不同的心脏细胞,即第一心室与第二心室(Bruneau,2008;De Pater et al.,2009)。miR-1是第一个被发现的心脏发育特异miRNA,在心脏发育中有着至关重要的作用(Zhao et al.,2005)。miRNA参与调控心室的移动,如miR-218a-1/2通过内皮生长因子调控心内膜迁移(Fish et al.,2011)。
在斑马鱼中,第一心室产生的心管和第二心室细胞分别分化成平滑肌和心肌层,心管循环、收缩,形成心房和心室(Hami et al.,2011)。在斑马鱼心脏发育阶段敲除miR-138可导致心血管延伸受限和早期心室循环缺失;更进一步的研究发现,在房室管内miR-138通过调控视黄酸合成和直接抑制软骨素蛋白多糖2靶向房室管发育的基因(Morton et al.,2008)。miR-133与miR-143也参与鱼类心脏的发育,敲除或抑制miR-143能使心室塌陷及降低收缩力,miR-133调控心脏的再生(Deacon et al.,2010;Yin et al.,2012)。在斑马鱼心脏发育中,miR-218a与转录因子Tba5相互作用,这对心内膜细胞分化与瓣膜组织形成是必须的(Chiavacci et al.,2012)。miR-21通过调节萌芽同源体的表达和细胞程序性死亡及同源性磷酸酶B来调控心脏瓣膜形成(Banjo et al.,2013)。用不同浓度微囊藻毒素处理斑马鱼胚胎,结果发现微囊藻毒素导致miR-126表达下调,miR-126负调控血管内皮生长因子(VEGF)信号通路,敲除miR-126导致血管完整性缺失及出血(Zhao et al.,2011)。Lalwani等(2012)运用基因敲除和过表达的方法研究斑马鱼胚胎发育过程中的miRNA,发现miR-142-3p通过靶向cdh5而影响血管完整性、重建及血管再生。miR-144和miR-451在斑马鱼胚胎红细胞组织中特异表达(Su et al.,2014),miR-144通过抑制meis1而调节造血作用,miR-144过表达导致血红蛋白表达水平下降(Biyashev et al.,2012)。此外,let-7、miR-20b、miR-31、miR-221和miR-181a在斑马鱼胚胎发育过程中参与血管再生和淋巴管生成(Dore et al.,2008;Nicoli et al.,2012;Dunworth et al.,2013)。
2.6 miRNA在鱼类骨骼形成中的作用鱼类骨骼形成包括软骨和硬骨的形成,被几种常见、特异的基因调控。骨骼的主要部分起源于神经嵴、侧向中胚层、近轴中胚层和脊索。间质细胞依赖转录因子和信号途径形成软骨细胞、成骨细胞和其他骨骼细胞(Karsenty & Wagner,2002)。在斑马鱼软骨分化中,miR-140通过调控转录因子sox9特异表达调控其发育,还通过调节血小板衍生生长因子受体α调控上颚骨骼发育。有研究发现,miR-140过表达导致斑马鱼发育中形成唇裂和腭裂(Eberhart et al.,2008)。Runx2也是调控成骨分化的转录因子,Huang等(2010)研究表明,在基质与成肌细胞间miR-204/miR-211直接负调控Runx2。以上结果说明,miRNA在骨骼与软骨组织形成过程中发挥重要作用。
2.7 miRNA在鱼类其他发育中的作用在鱼类发育过程中,miR-203和miR-133参与调控鳍再生。在斑马鱼鳍再生过程中,miR-203直接靶向Wnt信号途径中的转录因子Lef1,使鳍再生受到阻遏;而敲除miR-203使Lef1的表达水平增加,导致鳍过度生长(Thatcher et al.,2008)。miR-203也是一种皮肤特异miRNA,在牙鲆变态发育中,miR-203和miR-25分别调控皮肤发育和色素形成(Mitsuo et al.,2010)。miR-30和miR-429在尼罗河罗非鱼的肾和鳃中高表达,同时调节鱼体内渗透压(Yan et al.,2012b)。miR-10通过抑制HoxB1a和HoxB3a表达而调节斑马鱼发育中前后轴的发育,敲除miR-10导致HoxB1a和HoxB3a表达水平上升,而过表达miR-10则导致HoxB1a和HoxB3a功能缺失(Woltering & Durston,2008)。
3 结语与展望近年来,miRNA已成为生命科学领域一个研究热点。大多数研究主要探讨miRNA的表达及鉴定,其功能研究尚不充分和深入。miRNA参与生物体的生长发育、细胞增殖、分化及凋亡等多种生理和病理过程。miRNA对生物体胚胎发育过程中组织器官的形成具有重要的调控作用。鱼类作为重要的脊椎动物,在生物学研究领域占有重要地位,目前鱼类miRNA研究主要集中在miRNA的表达鉴定及新miRNA发现,而对鱼类miRNA的功能及其作用的分子机制知之甚少。近年来,多项研究报道了miRNA在鱼类胚胎发育过程中的调控作用,了解miRNA在鱼类胚胎发育中的功能和分子机制,可以为鱼类生长发育、繁殖及疾病防御提供良好的理论基础。同时,也可使我们对脊椎动物研究的生物学途径有更全面的了解,并有助于深入理解miRNA在人类基因中的调控网络。
致谢: 衷心感谢导师李效宇教授的悉心指导,并提供了宝贵的参考意见和建议。
| 丁雷, 同学春, 孙效文, 等. 2011. MicroRNAs对斑马鱼发育的调控[J]. 遗传, 33(11): 1179-1184. |
| 郝克红, 段涛, 王凯, 等. 2011. MicroRNA的研究进展[J]. 医学综述, 17(19): 2884-2887. |
| 唐雪莲, 李洪, 付京花. 2013. 鱼类microRNAs研究进展[J]. 水产科学, 32(1): 55-58. |
| 王佳佳, 徐超, 屠云杰, 等. 2007. 斑马鱼及其胚胎在毒理学中的实验研究与应用进展[J]. 生态毒理学报, 2(2): 123-135. |
| 张满仓, 吕艳, 戚艳婷, 等. 2008. 反义抑制和过表达miR-219引起斑马鱼胚胎发育异常[J]. 分子细胞生物学报, 41(5): 342-348. |
| Ambros V. 2004. The functions of animal microRNAs[J]. Nature, 431(7006): 350-355. |
| Banjo T, Grajcarek J, Yoshino D, et al. 2013. Haemodynamically dependent valvulogenesis of zebrafish heart is mediated by flow-dependent expression of miR-21[J]. Nature Communications, 4: 1978. |
| Bartel DP. 2004. MicroRNAs:genomics, biogenesis,mechanism, and function[J]. Cell, 116: 281-297. |
| Bekaert M, Natalie RL, Stephen CB, et al. 2013. Sequencing and characterisation of an extensive Atiantic salmo (Salmo salar L.) microRNA repertoire[J]. PLoS One, 8(7): e70136. |
| Betel D, Wilson M, Gabow A, et al. 2008. The microRNA.org resource:targets and expression[J]. Nucleic Acids Research, 36: D149-D153. |
| Biyashev D, Veliceasa D, Topczewski J, et al. 2012. miR-27b controls venous specification and tip cell fate[J]. Blood, 119: 2679-2687. |
| Bobe J, Andre S, Fauconneau B. 2000. Embryonic muscle development in rainbow trout (Oncorhynchus mykiss): a scanning electron microscopy and immunohistological study[J]. Journal of Experimental Zoology, 286(4): 379-389. |
| Borchert GM, Lanier W, Davidson BL. 2006. RNA polymerase Ⅲ transcribes human micro RNAs[J]. Nature Structural & Molecular Biology, 13: 1097-1101. |
| Bruneau BG. 2008. The development algenetics of congenital of congenital heart disease[J]. Nature, 451: 943-948. |
| Cavodeassi F, Concha ML, Houart C, et al. 2005. Early stages of zebrafish eye formation require the coordinated activity of Wnt11, Fz5, and the Wnt/b-catenin pathway[J]. Neuron, 47: 43-56. |
| Chi W, Tong C, Gan X, et al. 2011. Characterization and comparative profiling of miRNA transcriptomes in bighead carp and silver carp[J]. PLoS One, 6(8): e23549. |
| Chiavacci E, Dolfi L, Verduci L, et al. 2012. MicroRNA-218 mediates the effects of Tbx5a overexpression on zebrafish heart development[J]. PLoS One, 7: e50536. |
| Conte I, Carrella S, Avellino R, et al. 2010. miR-204 is required for lens and retinal development via Meis2 targeting[J]. Proceedings of the National Academy of Sciences, 107: 15491-15496. |
| Davis BN, Hata A. 2009. Regulation of microRNA biogenesis: a myriad of mechanisms[J]. Cell Communication and Signaling, 7: 18. |
| De Pater E, Clijsters L, Marques SR, et al. 2009. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart[J]. Development, 136: 1633-1641. |
| Deacon DC, Nevis KR, Cashman TJ, et al. 2010. The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis[J]. Development, 137(11): 1887-1189. |
| Dore LC, Amigo JD, Dos Santos CO, et al. 2008. A GATA-1-regulated microRNA locus essential for erythropoiesis[J]. Proceedings of the National Academy of Sciences, 105: 3333-3338. |
| Dunworth WP, Cagavi E, Kim JD, et al. 2013. Bone morphogenetic protein 2 signaling negatively modulates lymphatic development in vertebrate embryos[J]. Circulation Research, 114: 56-66. |
| Eberhart JK, He X, Swartz ME, et al. 2008. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis[J]. Nature Genetics, 40: 290-298. |
| Fang Z, Rajewsky N. 2011. The impact of miRNA target sites in coding sequences and in 3'UTRs[J]. PLoS One, 6(3): e18067. |
| Fish JE, Wythe JD, Xiao T, et al. 2011. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish[J]. Development, 138: 1409-1419. |
| Flynt AS, Li N, Thatcher EJ, et al. 2007. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate[J]. Nature Genetics, 39(2): 259-263. |
| Fu Y, Shi Z, Wu M, et al. 2011. Identification and differential expression of microRNAs during metamorphosis of the Japanese flounder (Paralichthys olivaceus)[J]. PLoS One, 6(7): 22957. |
| Gestri G, Link BA, Neuhauss S. 2012. The visual system of zebrafish and its use to model human ocular diseases[J]. Developmental Neurobiology, 72: 302-327. |
| Giraldez AJ, Cinalli RM, Glasner ME, et al. 2005. MicroRNAs regulate brain morphogenesis in zebrafish[J]. Science, 308: 833-838. |
| Giraldez AJ, Mishima Y, Rihel J, et al. 2006. Zebrafish miR-430 promotes deadenylation and clearance of maternal mRNAs[J]. Science, 312: 75-79. |
| Hami D, Grimes AC, Tsai HJ, et al. 2011. Zebrafish cardiac development requires a conserved secondary heart field[J]. Development, 138: 2389-2398. |
| He L, Hannon GJ. 2004. MicroRNAs: small RNAs with a big role in gene regulation[J]. Nature Reviews Genetics, 5: 522-531. |
| Huang J, Zhao L, Xing L, et al. 2010. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation[J]. Stem Cells, 28: 357-364. |
| IUCN. 2011. IUCN Red List of Threatened Species[EB/OL]. Version 2011. 2. [2012-03-02]. http://www.iucnredlist.org/. |
| Johansen SD, Karlsen BO, Furmanek T, et al. 2011. RNA deep sequencing of the Atlantic cod transcriptome[J]. Comparative Biochemistry and Physiology-Part D: Genomics and Proteomics, 6(1): 18-22. |
| Johnston IA. 2006. Environment and plasticity of myogenesis in teleost fish[J]. Journal of Experimental Biology, 209: 2249-2264. |
| Kapralova KH, Franzdottir SR, Jonsson H, et al. 2014. Patterns of miRNA expression in Arctic charr development[J]. PLoS One, 9(8): e106084. |
| Kapsimali M, Kloosterman WP, Rosa F, et al. 2007. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system[J]. Genome Biology, 8: R173. |
| Karsenty G, Wagner EF. 2002. Reaching a genetic and molecular understanding of skeletal development[J]. Developmental Cell, 2: 389-406. |
| Kerr TA, Korenblat KM, Davidson NO. 2011. MicroRNAs and liver disease[J]. Translational Research, 157(4): 241-252. |
| Ketley A, Warren A, Holmes E, et al. 2013. The miR-30 MicroRNA family targets smoothened to regulate Hedgehog signaling in zebrafish early muscle development[J]. PLoS One, 8(6): e65170. |
| Lakshmipathy U, Hart RP. 2008. Concise Review: microRNA expression in multipotent mesenchymal stromal cells[J]. Stem Cells, 26(2): 356-363. |
| Lalwani MK, Sharma M, Singh AR, et al. 2012. Reverse genetics screen in zebrafish identifies a role of miR-142a-3p in vascular development and integrity[J]. PLoS One, 7: e52588. |
| Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14[J]. Cell, 75: 843-854. |
| Lee Y, Ahn C, Han J, et al. 2003. The nuclear RNase Ⅲ Drosha initiates microRNA processing[J]. Nature, 425: 415-419. |
| Leucht C, Stigloher C, Wizenmann A, et al. 2008. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary[J]. Nature Neuroscience, 11(6): 641-648. |
| Lewis BP, Bartel DP, Burge CB, et al. 2003. Prediction of mammalian microRNA targets[J]. Cell, 115(7): 787-798. |
| Li SC, Chan WC, Ho MR, et al. 2010. Discovery and characterization of medaka miRNAs genes by next generation sequencing platform[J]. BMC Genomics, 11(Suppl 4): 8. |
| Lim B, Teh C, Shyh-Chang N, et al. 2009. MicroRNA-125b is a novel negative regulator of p53[J]. Genes Development, 23(7): 862-876. |
| Liu X, Ning G, Meng A, et al. 2012. MicroRNA-206 regulates cell movements during zebrafish gastrulation by targeting prickle1a and regulating c-Jun N-terminal kinase 2 phosphorylation[J]. Molecular and Cellular Biology, 32: 2934-2942. |
| Ma H, Hostuttler M, Wei H, et al. 2012. Characterization of the rainbow trout egg microRNA transcriptome[J]. PLoS One, 7(6): e39649. |
| Memczak S, Jens M, Elefsinioti A, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency[J]. Nature, 495: 333-338. |
| Mishima Y, Abreu-Goodger C, Staton AA, et al. 2009. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization[J]. Genes & Development, 23: 619-632. |
| Mitsuo N, Tadahisa S, Masato A, et al. 2010. Dual appearance of xanthophores, and ontogenetic changes in other pigment cells during early development of Japanese flounder Paralichthys olivaceus[J]. Fisheries Science (Tokyo), 76: 243-250. |
| Morton SU, Scherz PJ, Cordes KR, et al. 2008. MicroRNA-138 modulates cardiac patterning during embryonic development[J]. Proceedings of the National Academy of Sciences, 105: 17830-17835. |
| Nicoli S, Knyphausen CP, Lakshmanan A, et al. 2012. MiR-221 is required for endothelial tip cell behaviors during vascular development[J]. Developmental Cell, 22: 418-429. |
| Pasquinelli AE, Reinhart BJ, Shck FJ, et al. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA[J]. Nature, 408(6808): 86-89. |
| Pierce ML, Weston MD, Fritzsch B, et al. 2008. MicroRNA-183 family conservation and ciliated neurosensory organ expression[J]. Evolution & Development, 10: 106-113. |
| Pillai RS, Bhattacharyya SN, Filipowicz W. 2007. Repression of protein synthesis by miRNAs: how many mechanisms?[J]. Trends in Cell Biology, 17: 118-126. |
| Ramachandran R, Fausett BV, Goldman D. 2010. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signaling pathway[J]. Nature Cell Biology, 12: 1101-1107. |
| Reinhart BJ, Slack FJ, Basson M, et al. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans[J]. Nature, 403: 901-906. |
| Salem M, Xiao C, Womack J, et al. 2010. A microRNA repertoire for functional genome research in rainbow trout (Oncorhnchus mykiss)[J]. Marine Biotechnology (NY), 12(4): 410-429. |
| Shrivastava S, Steele R, Ray R, et al. 2015. MicroRNAs: role in hepatitis C virus pathogenesis[J]. Gene & Diseases, 2(1): 35-45. |
| Song L, Tuan RS. 2006. MicroRNAs and cell differentiation in mammalian development[J]. Birth Defects Research Part C: Embryo Today, 78(2): 140-149. |
| Soni K, Choudhary A, Patowary A, et al. 2013. MiR-34 is maternally inherited in Drosophila melanogaster and Danio rerio[J]. Nucleic Acids Research, 41: 4470-4480. |
| Su Z, Si W, Li L, et al. 2014. MiR-144 regulates hematopoiesis and vascular development by targeting meis1 during zebrafish development[J]. The International Journal of Biochemistry & Cell Biology, 49: 53-63. |
| Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts[J]. Development, 136: 3303-3042. |
| Tani S, Kusakabe R, Naruse K, et al. 2010. Genomic organization and embryonic expression of miR-430 in medaka (Oryzias latipes): insights into the post-transcriptional gene regulation in early development[J]. Gene, 449(1/2): 41-49. |
| Thatcher EJ, Paydar I, Anderson KK, et al. 2008. Regulation of zebrafish fin regeneration by microRNAs[J]. Proceedings of the National Academy of Sciences, 105: 18384-18389. |
| Townley-Tilson WH, Callis TE, Wang D. 2010. MicroRNAs 1, 133 and 206: critical factors of skeletal and cardiac muscle development, function,and disease[J]. The International Journal of Biochemistry & Cell Biology, 42: 1252-1255. |
| Wang X, Ono Y, Tan S, et al. 2011. Prdm1a and miR-499 act sequentially to restrict Sox6 activity to the fast-twitch muscle lineage in the zebrafish embryo[J]. Development, 138: 4399-4404. |
| Wienholds E, Kloosterman WP, Miska E, et al. 2005. MicroRNA expression in zebrafish embryonic development[J]. Science, 309: 310-311. |
| Wienholds E, Koudijs MJ, Cuppen E, et al. 2003. The microRNA-producing enzyme Dicer1 is essential for zebrafish development[J]. Nature Genetics, 35: 217-218. |
| Wightman B, Ha I, Ruvkun G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C.elegans[J]. Cell, 75: 855-862. |
| Woltering JM, Durston AJ. 2008. MiR-10 represses HoxB1a and HoxB3a in zebrafish[J]. PLoS One, 3(1): e1396. |
| Xia J, He X, Bai Z, et al. 2011. Identification and characterization of 63 microRNAs in the Asian seabass Lates calcarifer[J]. PLoS One, 6: 11. |
| Xie C, Xu S, Yang L, et al. 2011. mRNA/microRNA profile at the metamorphic stage of olive flounder (Paralichthys olivaceus)[J]. Comparative and Functional Genomics, 2011: 256038. |
| Xu S. 2009. MicroRNA expression in the eyes and their significance in relation to functions[J]. Progress in Retinal and Eye Research, 28: 87-116. |
| Xu Z, Chen J, Li X, et al. 2013. Identification and characterization of microRNAs in channel catfish (Ictalurus punctatus) by using solexa sequencing technology[J]. PLoS One, 8(1): e54174. |
| Xu Z, Qin Q, Ge J, et al. 2012. Bioinformatic identification and validation of conservative microRNAs in Ictalurus punctatus[J]. Molecular Biology Reports, 39: 10395-10405. |
| Yan B, Guo JT, Zhao LH, et al. 2012a. MicroRNA expression signature in skeletal muscle of Nile tilapia[J]. Aquaculture, 364-365: 240-246. |
| Yan B, Guo JT, Zhao LH, et al. 2012b. MiR-30c: a novel regulator of salt tolerance in tilapia[J]. Biochemical and Biophysical Research Communications, 425: 315-320. |
| Yan B, Zhu CD, Guo JT, et al. 2013. miR-206 regulates the growth of the teleost tilapia (Oreochromis niloticus) through the modulation of IGF-1 gene expression[J]. Journal of Experimental Biology, 216: 1265-1269. |
| Yin VP, Lepilina A, Smith A, et al. 2012. Regulation of zebrafish heart regeneration by miR-133[J]. Developmental Biology, 365: 319-327. |
| Zhao Y, Samal E, Srivastava D. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis[J]. Nature, 436: 214-220. |
| Zhao Y, Xiong Q, Xie P. 2011. Analysis of microRNA expression in embryonic developmental toxicity induced by MC-RR[J]. PLoS One, 6 (7): e22676. |
| Zhu Y, Xue W, Wang J, et al. 2012. Identification of common carp (Cyprinus carpio) microRNAs and microRNA-related SNPs[J]. BMC Genomics, 13: 413. |
 2015, Vol.
2015, Vol.