文章信息
- 上官凌飞, 王子诚, 方项, 陈春, 刘天华, 陈梦霞, 房经贵
- SHANGGUAN Lingfei, WANG Zicheng, FANG Xiang, CHEN Chun, LIU Tianhua, CHEN Mengxia, FANG Jinggui
- DAM基因调控多年生果树芽休眠的机制
- Regulation mechanism of bud dormancy by DAM gene in perennial fruit trees
- 南京农业大学学报, 2020, 43(5): 790-799
- Journal of Nanjing Agricultural University, 2020, 43(5): 790-799.
- http://dx.doi.org/10.7685/jnau.201912069
-
文章历史
- 收稿日期: 2019-12-30
休眠是植物在进化过程中形成的一种对环境和季节性气候变化的生物学适应性机制[1], 也被称为“生命的隐蔽现象”, 对物种的生存和繁衍具有重要的生物学和生态学意义, 在农业生产上还具有重要的经济意义[2]。
Lang等[3]将休眠定义为“包括分生组织在内的任何结构可见生长的暂时停止”, 并根据休眠的诱导因素将其分为3类:1)类休眠(paradormancy), 指休眠结构(组织或器官)以外的生理因素(如顶端优势、激素等)诱导产生的生长停滞现象; 2)内休眠(endodormancy), 指休眠结构(组织或器官)本身的因素(如冷温需求、光周期影响)控制的生长停滞现象; 3)生态休眠(ecodormancy), 是指由环境因子(如温度、水分胁迫、营养亏缺等)引起的休眠, 一旦环境条件适宜, 植物便可重新恢复生长。芽休眠是高等植物经过长期演化而获得的一种对极端环境条件和季节性变化相适应的有益的生物学特性, 因此, 芽休眠也是一种对逆境环境(如低温)的应答机制。一般来说, 影响休眠的因素包括树种与品种[4]、外界因素(光照、温度、水分)[5-7]、内部因素(激素、糖、酶)[8-11]、休眠相关基因[PHY(Phytochrome)、CBF(C-repeat binding factor)、CYC(Cyclin)、DAM、SVP(Short vegetative phase)等][12-17]、表观遗传调控机制[18-19]等。
DAM(Dormancy-associated MADS-box)基因是近十年来发现的与芽休眠调控关系密切相关的基因之一。笔者总结了近几年DAM基因鉴定与功能分析, 发现这类基因对模式植物和多年生果树生长发育起不同的调控作用, 说明这些基因在进化过程中形成多样化的功能。另外本文重点介绍DAM基因调控多年生果树芽休眠机制以及影响DAM基因表达的因子, 以期为深入研究DAM基因调控多年生果树芽休眠解除机制提供参考和借鉴。
1 DAM基因鉴定与功能SVP(Short vegetative phase, 登录号:AT2G22540)和AGL 24(Agamous-like 24, 登录号:AT4G24540)是拟南芥中被证实的2个参与成花途径调控的关键基因, 能够直接调控FT(Flowering locus T)基因的表达来影响成花途径[20]。Bielenberg等[14]从桃突变体evergrowing(evg)中发现6个串联重复的DAM基因位于evg位点, 其中PpDAM5和PpDAM6在突变体中表达下调。系统进化分析显示, DAM基因与拟南芥SVP和AGL 24基因最为相似。随着对DAM和SVP基因研究的不断深入, DAM与SVP基因被证实能够影响种子萌发与花形态建成, 如超表达樱桃PpcDAM6基因的拟南芥种子萌发明显受到抑制[21]; 白梨PpDAM1蛋白能够通过结合在PpFT 2基因启动子区域来抑制其表达[22]; 异源表达樱桃PavDAM 1和PavDAM5会导致转基因拟南芥花发育异常[23]。除此之外, 更多的研究报道显示, DAM基因的另一重要功能便是调控芽休眠进程[22, 24-26]。
近十年来, 在桃、梨、蓝莓、樱桃、梅、苹果、猕猴桃等多个物种上都发现DAM基因的存在, 且不同物种间DAM基因的数量呈现一定的差异。由表 1可知:多数DAM基因在不同物种中能够直接或间接调控芽休眠进程, 对芽生长产生一定的抑制作用, 并且不同物种DAM基因, 甚至同物种不同DAM类基因的作用都不同, 说明DAM基因既有保守性又有多样性。
| 物种 Species |
DAM基因数 Number of DAM genes |
基因名 Gene name |
登录号 Accession number |
基因功能 Gene function |
参考文献 References |
| 樱桃 Cherry |
3 | PpcDAM4 | KM243365 | 与植株生长发育及休眠有关* Related to plant growth and dormancy* |
[27] |
| PpcDAM5 | KM243366 | 参与芽休眠及休眠解除进程* Participating in bud dormancy and dormancy release process* |
[27] | ||
| PpcDAM6 | KM243367 | 抑制种子萌发* Inhibiting seed germination* |
[21] | ||
| 蓝莓 Blueberry |
1 | VcDAM1 | KF871069 | 参与蓝莓花芽休眠进程 Participating in the process of blueberry flower bud dormancy |
[28] |
| 梨 Pear |
4 | PpMADS13-2 | AB504717 | 可能参与芽休眠调控及开花过程中花器官的发育* May be involved in regulation of bud dormancy and development of flower organs during flowering* |
[29-30] |
| PpMADS13-3 | AB774474 | 可能参与芽休眠调控及开花过程中花器官的发育* May be involved in regulation of bud dormancy and development of flower organs during flowering* |
[29-30] | ||
| PpyMADS31 | 103964948 | 可能参与芽休眠调控及开花过程中花器官的发育* May be involved in regulation of bud dormancy and development of flower organs during flowering* |
[29] | ||
| PpMADS13-l | AB504716 | 可能参与芽休眠调控* May be involved in the regulation of bud dormancy* |
[29-30] | ||
| 桃 Peach |
2 | PpDAM5 | DQ863251 | 抑制芽生长 Inhibiting bud growth |
[31-32] |
| PpDAM6 | DQ863252 | 抑制芽生长 Inhibiting bud growth |
[16, 31-32] | ||
| 梅 Plum |
2 | PmDAM5 | AB576349 | 转录抑制因子, 影响花蕾的生长和休眠 Transcriptional suppressor, affecting bud growth and dormancy |
[33-34] |
| PmDAM6 | AB437345 | 过表达抑制芽生长, 影响花蕾的生长和休眠* Overexpression inhibits bud growth and affects bud growth and dormancy* |
[33-34] | ||
| 苹果 Apple |
6 | MdDAMb | ADL36743 | 过表达延迟萌芽, 减少侧枝生长 Overexpression delays germination and reduces lateral branch growth |
[35-36] |
| MdDAM1 | KP164996 | 参与芽休眠, 开花抑制因子 Participating in bud dormancy, flowering inhibition factor |
[35-36] | ||
| MdDAM2 | KP164997 | 参与芽休眠 Participating in bud dormancy |
[35] | ||
| MdDAM4 | KT582789 | 参与芽休眠 Participating in bud dormancy |
[35] | ||
| MdDAM3 | LC004730 | 开花抑制因子 Flowering inhibition factor |
[35] | ||
| MdSVPa | KT582788 | 过表达延迟萌芽, 减少侧枝生长 Overexpression delays germination and reduces lateral branch growth |
[24, 35] | ||
| 猕猴桃 Kiwifruit |
4 | AcSVP1 | JF838212 | 延迟开花, 影响花器官发育 Delaying flowering and affecting the development of floral organs |
[26] |
| AcSVP2 | JF838213 | 抑制分生组织的生长, 延迟萌芽 Inhibiting the growth of meristematic tissue, delaying germination |
[37] | ||
| AcSVP3 | JF838214 | 延迟开花, 影响花器官、果实及种子发育 Delaying flowering and affecting flower organs, fruit and seed development |
[38] | ||
| AcSVP4 | JF838215 | 参与芽休眠及开花调控 Participating in bud dormancy and flowering regulation |
[26] | ||
| 杏 Apricot |
2 | ParDAM5 | MH464454 | 延迟休眠解除及开花时间 Delaying dormancy release and flowering time |
[39] |
| ParDAM6 | MH464455 | 延迟休眠解除及开花时间 Delaying dormancy release and flowering time |
[39] | ||
| 葡萄 Grape |
5 | VvSVP1 | VIT_00s0313g00070 | 抑制花组织发育* Inhibiting floral tissue development |
[40] |
| VvSVP2 | VIT_18s0001g07460 | 可能与芽的发育有关* May be related to bud development |
[40] | ||
| VvSVP3 | VIT_15s0107g00120 | 抑制花组织发育* Inhibiting floral tissue development |
[40] | ||
| VvSVP4 | VIT_03s0017g00360 | 抑制花组织发育* Inhibiting floral tissue development |
[40] | ||
| VvSVP5 | VIT_03s0167g00100 | 可能与芽的发育有关* May be related to bud development |
[40] | ||
| 注:*表示该功能为预测, 其余则表示功能鉴定。 Note:*indicates the predicated function, and the function of the remaining genes is verified. |
|||||
在苹果中过表达MdDAMb和MdSVPa能够推迟芽的萌发并且减少侧枝的生长[24]。在拟南芥中发现介导低温响应途径的CBF基因与CRT/DRE(C-repeat/dehydration responsive element)调控元件结合, 能激活调控与低温有关的基因表达[41]。另有研究发现超表达PpcDAM 4可抑制樱桃的生长发育[27], 可能是PpcDAM 4 启动子序列中存在与光调控、响应低温顺式作用的DNA调控元件(CRT/DRE), 从而调控植株生长进程。Prassinos等[42]在甜樱桃上发现与DAM 3相似性非常高的基因DAM3 -like, 而且矮化型相对于非矮化型该基因表达上调, 表明DAM 3 -like基因能抑制植株生长, 从而达到矮化植株的目的。在高需冷量猕猴桃中过表达AdSVP 2会推迟芽的萌发, AdSVP2可以抑制分生组织的生长, 从而抑制芽在满足低温需冷量之前的萌发, 但长期暴露在低温中可解除这种抑制效果。但在低需冷量猕猴桃中, 过表达AdSVP 2则没有类似的效果[37]。另外, Diaz-Riquelme等[40]在对葡萄MIKC基因研究中发现, VvSVP 1、VvSVP3、VvSVP4主要在芽内表达, 且可抑制休眠前花芽分生组织的生长。以上研究表明DAM基因能够抑制芽生长, 但其调控的下游途径还未知, 需要利用分子生物学手段进行进一步挖掘与分析。
2.2 DAM基因对FT类基因的抑制作用FT类基因被证实与温带树木芽休眠相关, 能够改变植株生长, 使其产生过早开花等表型[43]。桑树FLC-like基因MaMADS 33在内休眠进程中的表达模式与DAM类似, 该基因可直接或间接结合到MaFT基因启动子区域, 抑制下游报告基因的表达, 从而调控芽休眠[44]。白梨PpDAM1蛋白可通过结合PpFT 2基因启动子区域来抑制其表达, 且PpFT2基因在休眠期表达水平低, 而休眠解除期表达水平高, 与PpDAM基因正好相反[22]。SVP是在营养阶段调节猕猴桃[26]和葡萄[45]的FT、SOC 1 (Supressor of overexpression of constans 1)和FLC表达的开花抑制基因。Busov[46]在拟南芥中发现SVL可直接结合并激活TCP8/BRANCHED1蛋白的表达, TCP8/BRANCHED1蛋白可进一步抑制FT基因表达。高真真等[47]在油桃芽休眠研究中发现, PpSVP-like基因在自然休眠与休眠解除期表达量逐渐下降, 在花芽休眠解除期达到最低值, 而PpFT-like基因在这一过程中表达持续上调, 说明PpSVP-like基因和PpFT-like基因对芽休眠及其解除有相反的作用, 推测SVP-like基因可通过某种机制来抑制FT-like基因的表达。但是, 研究人员在日本梨和猕猴桃上没有证实FT-like基因是PpMADS 13-1(PpDAM1)和AcSVP2的靶基因[48-49]。因此, 对于DAM/SVP-like(SVL)与FT-like基因之间的调控模式还有待进一步研究。
2.3 DAM基因与植物激素之间的相互调控SVL被认为至少部分通过转录激活脱落酸(ABA)合成限速酶编码基因NCED 3 (9-cis-epoxycarotenoid dioxygenase 3)和ABA受体基因来抑制杂交杨的萌芽, 外源施加ABA可以促进杨树SVL基因的表达[17]。研究表明, 在日本梨中PpDAM 1 通过结合PpNCED 3 启动子区域的顺式元件(CArG box)激活其转录, 而NCED的表达水平决定了植物体内ABA的含量。ABA下游响应基因又会在内生性休眠解除过程中下调PpDAM 1 表达, ABA和DAM之间存在着反馈调节机制[50]。Yamane等[51]研究表明, 梅的PmDAM 6 基因可通过调节休眠芽中ABA和细胞分裂素(CTK)的含量来抑制芽萌发。另外, 尽管在猕猴桃中AcSVP2能够结合到ABA、渗透胁迫以及干旱胁迫等响应基因上, 却没有发现AcSVP2能够结合到NCED同源基因启动子区域[49]。这些研究结果说明DAM/SVP蛋白介导ABA调控芽休眠进程可能存在多种机制。研究表明, 苹果MdSVP同源基因MdMADS 50 启动子中包含1个ABA响应元件ABRE(ABA-responsive element), 推测MdMADS 50 可能参与了干旱调节途径[52]。除了ABA, 对草莓进行赤霉素处理, FaSVP 1 在解除休眠阶段表达量持续下降[53]。在杂交杨[17]和拟南芥[54]中, SVL也能够抑制GA合成酶编码基因GA 20ox(Gibberellin 20 oxidase)的表达水平, 从而抑制花芽分化。但DAM/SVL基因如何通过调控GA水平来参与果树芽休眠进程尚不明确。
3 DAM基因表达的影响因子 3.1 转录因子在梨[22]、苹果[55]和梅[56]中DAM基因的启动子区域均含有CBF转录因子的调控结合元件CRT/DRE, 通过酵母单杂交试验发现CBF蛋白能够分别与梨PpDAM 1和PpDAM3 [22]以及梅PmDAM 6 基因启动子区域[56]相结合。CBF能够激活DAM基因的表达, 且转CBF基因植株表现出抗寒性增强、生长暂时停止和叶片衰老提前以及破芽延迟等表型[13, 22, 48, 55]。PpAREB1和PpHB22能够结合到PpDAM 1 的启动子区域, 并调控其表达[50, 57]。Wang等[58]在桃‘中油4’品种中分离出了Teosinte branched1/cycloidea/proliferating cell factor(TCP)转录因子PpTCP20, 在芽休眠解除过程中PpTCP20可结合到PpDAM 5和PpDAM6基因的启动子区域, 阻止基因表达, 进而抑制芽休眠。以上多个物种上分子互作试验表明, CBF、AREB以及HB等转录因子能够有效结合DAM基因启动子区域, 进而调控DAM基因表达。
3.2 表观遗传机制Leida等[19]在研究大戟休眠芽时提出DAM基因的表观遗传调控, 并在EesDAM 1 转录起始位点下游的2个区域发现H3K4me3(组蛋白H3赖氨酸第4位上的3甲基化)水平降低和H3K27me3(组蛋白H3赖氨酸第27位上的3甲基化)富集的现象。de la Fuente等[59]发现桃DAM 6 表达水平在休眠解除中下调, 伴随其启动子区域、转录起始位点区域以及最大内含子区域H3ac和H3K4me3表达水平降低以及H3K27me3富集, 通过Chip-seq进一步发现在休眠解除芽中桃DAM 1和DAM4-6 基因也存在H3K27me3修饰水平显著富集。在梨中, DAM同源基因PpyMADS 13-1在内生性休眠解除前H3K4me3表达水平降低, 而H3K27me3表达水平却没有变化[48]。在梨休眠解除过程中, 组蛋白H2A.Z在PpyDAM 1 的TSS区域中的比例在休眠解除后下调, 暗示H2A.Z可能参与休眠解除过程中PpyDAM 1 的下调[48]。EAR(ERF-associated amphiphilic repression)结构域通过与一些共抑制子(如SAP18、SIN3等)互作, 最终与HDAC蛋白形成复合体, 对DNA结合区域的组蛋白去乙酰化, 通过染色体修饰途径达到抑制基因表达的作用[60]。由于EAR基序在桃[61]和梅[33] DAM蛋白中也被发现, 再结合前人对梨休眠相关基因的进化树结果分析[29], 推测出桃、梨DAM基因具有相同起源。因此, Kagale等[62]推测带有EAR结构域的梨PpyDAM蛋白可能通过EAR复合体去乙酰化途径抑制下游基因的表达, 调控休眠的建立与解除。在猕猴桃解除休眠过程中, H3K4蛋白三甲基化标记和H3蛋白乙酰化程度的降低导致AcSVP 2 表达量受到抑制, 进而解除芽休眠, 但是在AcSVP 2 位点上的H3K27和H3K9蛋白并没有在冬季休眠中有大量积累[63]。此外, microRNA(miRNA)也被证实参与DAM基因调控, miRNA6390能够靶向梨PpDAM, 以降解PpDAM从而释放PpFT, 促进内休眠的解除和芽的萌动[22]。表观遗传作为一种重要的研究手段, 在调控基因表达和生长发育方面起重要作用, 相信随着研究深入, 表观遗传机制将更好运用于研究果树芽休眠领域。
3.3 内含子区域在低积温需求桃中, DAM 5和DAM6第1个内含子区域都含有转座子相关的插入序列[64]。Porto等[35]发现在苹果、梅、梨和桃DAM基因内含子中同样存在一段高度保守序列。内含子区域通常比大部分DAM基因外显子序列要更加保守, 说明这类内含子在DAM转录调控过程中起功能性作用。da Silveira Falavigna等[65]研究表明, DAM内含子区域与AG(Agamous)、FLC(Flowering locus C)、FLM(Flowering locus M)等MADS-box基因结构特点相同, 推测内含子区域在进化过程中保留的这段保守序列可能会对DAM基因表达产生影响。
3.4 外源物质研究表明, 单氰胺(HC)处理促进桃芽休眠解除的同时, 还能下调PpDAM 5和PpDAM6的表达水平[31]。3个葡萄VvARP(Auxin-repressed protein)与DRMs(Dormancy-related proteins)基因在单氰胺处理中的表达量显著低于对照, 推测单氰胺处理使这些基因表达下调, 从而促进生长素的生物合成, 加速葡萄冬芽萌发进程[66]。对处于休眠期的蓝莓枝条喷施外源ABA, 蓝莓休眠芽萌发率明显降低, 并且VcDAM 1 基因上调表达, 推测外源ABA可通过促进DAM基因的表达来维持花芽休眠进程[28]。有研究发现, 调节细胞分裂素信号的ARR10 motif普遍存在于MdDAM基因启动子和整个内含子序列中, 推测细胞分裂素水平可间接抑制苹果DAM基因的表达[35]。GA和ABA被认为是芽休眠建立和解除过程中非常重要的2类激素。近年来关于这2类激素调控芽休眠的研究有较大进展, 但对休眠相关基因的分子调控机制尚不明确, 还有待深入研究。此外, 完善CTK、IAA等激素对芽休眠调控的分子网络也将是今后研究的主要方向。
3.5 温度通常认为低温能够打破休眠, 促进萌发。延长低温信号能下调SVP基因及其下游基因(TCP 18/BRC1和RCAR/PYL)的表达, 同时能上调FT1基因和GA途径相关基因的表达, 进而促进芽的萌发[29]。同样, SVL作为响应温度信号的内源介质可负调控芽休眠解除过程[46]。Niu等[22]研究发现, 梨芽经过短时低温能够诱导PpCBF的表达, 进而促进PpDAM基因的表达, 从而诱导内休眠的启动。另外, 作为ABA响应位点的ABI基因, 其表达可间接受到低温处理影响, 再进一步间接诱导梨DAM基因的表达[22]。人为延长低温处理时间, 桃树DAM 5和DAM6 基因表达量下调, 发芽率上升, 且发芽率与DAM 5、DAM6表达量呈负相关, 说明延长低温时间使桃树需冷量提前得到满足, 休眠提前解除, 同时伴随着DAM 5和DAM6基因表达下调[31]。研究表明, 高温处理可影响休眠相关基因表达量:梨花芽经过热处理后, PpMADS 13-1基因表达下调趋势比对照组明显, 且PpFT 2a基因表达提前上调, 最终导致花芽提前解除休眠[67]。
3.6 光周期多年生植物的周年生长特性决定其需要在不同的光照条件下完成整个生长发育过程。光周期往往通过影响细胞内某些物质的代谢来实现对芽休眠的调控。短日照条件下, 植物体内ABA大量积累, 而PKL(PICKLE, 一种染色质重塑因子)[68]和ABA与DAM/SVL均存在负调控关系, 推测短日照可通过这一途径间接调控DAM/SVL的表达。在梨PpyDAM的启动子区域还分布着多个其他顺式作用元件[22, 48], 这些顺式元件暗示PpyDAM也受光通路的调控[6]。Liu等[69]在双子叶植物中的SVP基因启动子区域已鉴定出23个光响应元件, 对这些元件的深入分析将会解释其如何影响SVP基因的表达及功能。
综上所述, DAM基因作为与芽休眠调控密切相关的基因之一, 其表达调控受多方面的因素影响, 它们共同组成了一个复杂的调控网络, 且随着研究的不断深入, 被不断完善(图 1)。
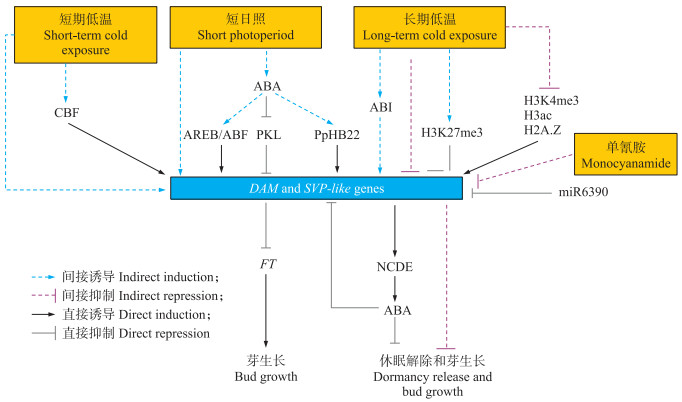
|
图 1 DAM和SVP-like基因调控的温带果树芽休眠分子调控网络模型[22, 65] Fig. 1 Molecular regulation network model of bud dormancy in temperate fruit trees regulated by DAM and SVP-like genes[22, 65] CBF:C重复结合因子C-repeat binding factor; ABA:脱落酸Abscisic acid; AREB/ABF:ABA response element binding protein/ABA binding factor; PKL:PICKLE; PpHB22:Homeodomain-leucine zipper(HD-Zip)protein; ABI:ABA-insensitive; H3K27me3:Histone H3 at lysine 27;H3K4me3:Histone H3 at lysine 4;H3ac:Acetylation levels of H3;H2A.Z:组蛋白Histone; NCDE:9-cis-epoxycarotenoid dioxygenase; FT:Flowering locus T. |
芽休眠是落叶果树栽培和设施果树生产的主要限制因素之一, 因此了解果树芽休眠调控机制显得尤为重要。从以上总结可看出近年来DAM基因调控的芽休眠分子机制网络模型得到了补充, 为后续研究提供了良好的参考方向。但由于芽休眠的形成与解除过程受到多种因素的影响, 仍存在某些调控途径尚不清晰的问题, 如在芽休眠调控过程中, 哪类基因在众多激素分子响应中起着主导作用[70]; 水分也是影响芽休眠的因素之一, 它在细胞中的状态与植株休眠存在一定相关性, 有研究发现休眠过程中高度表达的脱水蛋白可被ABA诱导, 但是其是否可作用于休眠相关基因来调控芽休眠还不清楚[71]。还有研究表明部分基因在芽休眠和种子休眠解除过程中表达水平一致[19], 推测二者可能有一些相似的调控网络, 可从这一角度来丰富芽休眠分子调控网络。随着分子生物学与基因组学的迅速发展, 综合利用基因组学、转录组学等技术手段来研究多种基因调控果树芽休眠诱导到解除的全过程, 将进一步完善芽休眠分子调控网络。
4.2 研究机制有助于破眠剂的开发研究芽休眠机制的最终目的是为生产提供有效的栽培管理技术, 提高经济效益。现实中由于生产上的需要, 往往要对植株进行破眠处理, 达到促进开花结果的目的。大量研究表明HC[31]、外源GA[53]等激素可解除果树芽休眠, 同时伴随DAM基因表达下调, 说明激素可作为破眠剂成分来达到解除果树休眠、促进萌发的目的。但是, 植物激素具体通过哪些通路来影响DAM基因表达还没有深入的研究。一定的高温处理可解除休眠, 这在树莓[72]、葡萄[40]等物种上已得到验证, 但高温如何作用于内休眠解除过程尚未明确, 因此对这一问题的研究将有助于解决果树秋季二次开花问题, 也会为新破眠技术的开发提供新的方向[6]。芽休眠机制研究在代谢组学上也有一定的进展, 员盎然等[73]研究推测通过调控丙氨酸解氨酶(PAL)、多酚氧化酶(PPO)、查尔酮异构酶(CHI)和肉桂酸-4-羟化酶(C4H)等关键酶的活性可控制酚类物质(杨梅酮、阿魏酸、表儿茶素等)含量的变化, 进而控制芽休眠与解除的进程。这些理论为后期深入研究芽休眠提供了新方向, 随着基因组学、转录组学、代谢组学的不断完善, 开发新型破眠剂前景广阔。
4.3 加强DAM基因功能研究目前对DAM调控机制研究主要集中在休眠的建立和解除方面:DAM基因表达受CBF、ARBE1等转录因子调控, 解释低温或短日照条件下芽休眠的建立; DAM也可通过调控FT、NCED等下游靶基因来参与休眠解除过程。但在休眠维持阶段, DAM基因如何表达以及与其他相关因子的互作关系还未可知, 亟待深入研究。转基因技术是基因功能鉴定的主要方法, 梨、樱桃等木本果树同源转基因工作较困难, 因此常以拟南芥为材料进行异源转基因功能验证, 通过观察种子萌发率和花形态建成、生理指标及相关基因表达等来推测基因功能。但部分DAM基因功能验证试验中转基因植株与对照植株种子萌发情况并无明显差异, 而过表达植株花发育往往出现异常[29, 44, 74]。虽然种子休眠与芽休眠具有一定相似性, 但该DAM基因对芽休眠是否有调控作用还有待进一步验证。因此, 在今后的研究中有必要加强果树转基因体系的研究, 利用同源转化体系来分析DAM基因的功能。此外, 鉴于DAM基因和激素在芽休眠中的重要作用以及芽休眠对多年生果树的重要性, 需要利用多组学联合分析技术结合酵母单/双杂交(Y1H/Y2H)、双分子荧光互补(BiFC)、凝胶迁移试验(EMSA)等技术系统挖掘DAM基因与激素协同调控多年生果树芽休眠的机制。
| [1] |
付立中, 申海林, 邹利人, 等. 落叶果树自然休眠研究进展[J]. 北方果树, 2014(4): 1-3. Fu L Z, Shen H L, Zou L R, et al. Research progress on natural dormancy of deciduous fruit trees[J]. Northern Fruits, 2014(4): 1-3 (in Chinese with English abstract). |
| [2] |
Samish R M. Dormancy in woody plants[J]. Annual Review of Plant Physiology, 1954, 5(1): 183-204. |
| [3] |
Lang G A, Early J D, Martin G C, et al. Endo-, para-, and ecodormancy:physiological terminology and classification for dormancy research[J]. Horticultural Science, 1987, 22: 371-377. |
| [4] |
王力荣, 朱更瑞, 方伟超, 等. 桃品种需冷量评价模式的探讨[J]. 园艺学报, 2003, 30(4): 379-383. Wang L R, Zhu G R, Fang W C, et al. Estimating models of the chilling requirement for peach[J]. Acta Horticulturae Sinica, 2003, 30(4): 379-383 (in Chinese with English abstract). |
| [5] |
Jian L C, Li P H, Sun L H, et al. Alterations in ultrastructure and subcellular localization of Ca2+ in poplar apical bud cells during the induction of dormancy[J]. Journal of Experimental Botany, 1997, 48(6): 1195-1207. DOI:10.1093/jxb/48.6.1195 |
| [6] |
Rohde A, Bhalerao R P. Plant dormancy in the perennial context[J]. Trends in Plant Science, 2007, 12(5): 217-223. DOI:10.1016/j.tplants.2007.03.012 |
| [7] |
Welling A, Palva E T. Molecular control of cold acclimation in trees[J]. Physiologia Plantarum, 2006, 127(2): 167-181. |
| [8] |
Le Mière P, Hadley P, Darby J, et al. The effect of temperature and photoperiod on the rate of flower initiation and the onset of dormancy in the strawberry(Fragaria×ananassa Duch.)[J]. Journal of Horticultural Science, 1996, 71(3): 361-371. |
| [9] |
Olsen J E, Junttila O, Moritz T. A localised decrease of GA1 in shoot tips of Salix pentandra seedings precedes cessation of shoot elongation under short photoperiod[J]. Physiologia Plantarum, 1995, 95(4): 627-632. DOI:10.1111/j.1399-3054.1995.tb05532.x |
| [10] |
Gibson S I. Control of plant development and gene expression by sugar signaling[J]. Current Opinion in Plant Biology, 2005, 8(1): 93-102. DOI:10.1016/j.pbi.2004.11.003 |
| [11] |
张昂.葡萄冬芽打破休眠过程中抗氧化机制及次生代谢的研究[D].杨凌: 西北农林科技大学, 2008. Zhang A. Research on antioxidant mechanism and secondary metabolism of grape winter buds during dormancy breaking[D]. Yangling: Northwest A & F University, 2008(in Chinese with English abstract). http://cdmd.cnki.com.cn/Article/CDMD-10712-2008102216.htm |
| [12] |
Kühn N, Ormeño-Núñez J, Jaque-Zamora G, et al. Photoperiod modifies the diurnal expression profile of VvPHYA and VvPHYB transcripts in field-grown grapevine leaves[J]. Journal of Plant Physiology, 2009, 166(11): 1172-1180. DOI:10.1016/j.jplph.2009.01.005 |
| [13] |
Wisniewski M, Norelli J, Bassett C, et al. Ectopic expression of a novel peach(Prunus persica)CBF transcription factor in apple(Malus×domestica)results in short-day induced dormancy and increased cold hardiness[J]. Planta, 2011, 233(5): 971-983. DOI:10.1007/s00425-011-1358-3 |
| [14] |
Bielenberg D G, Wang Y E, Li Z G, et al. Sequencing and annotation of the evergrowing locus in peach[Prunus persica(L.)Batsch]reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation[J]. Tree Genetics & Genomes, 2008, 4(3): 495-507. |
| [15] |
Wang S Y, Faust M. Changes in the antioxidant system associated with budbreak in 'Anna' apple(Malus×domestica Borkh.)buds[J]. Journal of the American Society for Horticultural Science, 1994, 119(4): 735-741. DOI:10.21273/JASHS.119.4.735 |
| [16] |
Yamane H, Kashiwa Y, Ooka T, et al. Suppression subtractive hybridization and differential screening reveals endodormancy-associated expression of an SVP/AGL24-type MADS-box gene in lateral vegetative buds of Japanese apricot[J]. Journal of the American Society for Horticultural Science, 2008, 133(5): 708-716. DOI:10.21273/JASHS.133.5.708 |
| [17] |
Singh R K, Miskolczi P, Maurya J P, et al. A tree ortholog of SHORT VEGETATIVE PHASE floral repressor mediates photoperiodic control of bud dormancy[J]. Current Biology, 2019, 29(1): 128-133. DOI:10.1016/j.cub.2018.11.006 |
| [18] |
Santamaría M E, Rodríguez R, Cañal M J, et al. Transcriptome analysis of chestnut(Castanea sativa)tree buds suggests a putative role for epigenetic control of bud dormancy[J]. Annals of Botany, 2011, 108(3): 485-498. |
| [19] |
Leida C, Conesa A, Llácer G, et al. Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner[J]. New Phytologist, 2012, 193(1): 67-80. DOI:10.1111/j.1469-8137.2011.03863.x |
| [20] |
Gregis V, Sessa A, Dorca-Fornell C, et al. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes[J]. The Plant Journal, 2009, 60(4): 626-637. DOI:10.1111/j.1365-313X.2009.03985.x |
| [21] |
梅忠, 朱友银, 刘向蕾, 等. 中国樱桃花芽休眠相关MADS-box基因的克隆与功能初探[J]. 植物生理学报, 2018, 54(9): 1433-1440. Mei Z, Zhu Y Y, Liu X L, et al. Isolation and functional analysis of dormancy-associated MADS-box gene in cherry(Prunus pseudocerasus)flower bud[J]. Plant Physiology Journal, 2018, 54(9): 1433-1440 (in Chinese with English abstract). |
| [22] |
Niu Q F, Li J, Cai D Y, et al. Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear(Pyrus pyrifolia white pear group)flower bud[J]. Journal of Experimental Botany, 2016, 67(1): 239-257. |
| [23] |
Wang J Y, Gao Z, Li H, et al. Dormancy-associated MADS-box(DAM)genes influence chilling requirement of sweet cherries and co-regulate flower development with SOC1 gene[J]. International Journal of Molecular Sciences, 2020, 21(3): 921. DOI:10.3390/ijms21030921 |
| [24] |
Wu R M, Tomes S, Karunairetnam S, et al. SVP-like MADS box genes control dormancy and budbreak in apple[J]. Frontiers in Plant Science, 2017, 8: 477. |
| [25] |
Bai S L, Saito T, Sakamoto D, et al. Transcriptome analysis of Japanese pear(Pyrus pyrifolia Nakai)flower buds transitioning through endodormancy[J]. Plant and Cell Physiology, 2013, 54(7): 1132-1151. DOI:10.1093/pcp/pct067 |
| [26] |
Wu R M, Walton E F, Richardson A C, et al. Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering[J]. Journal of Experimental Botany, 2012, 63(2): 797-807. |
| [27] |
邵姁.中国樱桃花芽休眠相关MADS-box转录因子克隆与功能分析[D].杭州: 浙江师范大学, 2016. Shao X. Cloning and functional analysis of MADS-box transcription factors related to dormancy of Chinese cherry flower buds[D]. Hangzhou: Zhejiang Normal University, 2016(in Chinese with English abstract). http://cdmd.cnki.com.cn/Article/CDMD-10345-1016289556.htm |
| [28] |
刘思, 夏秀英, 李波. 蓝莓花芽休眠相关MADS-box基因VcDAM1的克隆与表达分析[J]. 植物生理学报, 2017, 53(9): 1728-1734. Liu S, Xia X Y, Li B. Cloning and expression analysis of dormancy-associated MADS-box gene VcDAM1 from blueberry(Vaccinium corymbosum)[J]. Plant Physiology Journal, 2017, 53(9): 1728-1734 (in Chinese with English abstract). |
| [29] |
鄢馨卉.梨休眠相关DAM基因的鉴定及功能分析[D].杭州: 浙江大学, 2019. Yan X H. Identification and functional analysis of pear dormancy associated DAM genes[D]. Hangzhou: Zhejiang University, 2019(in Chinese with English abstract). http://cdmd.cnki.com.cn/Article/CDMD-10335-1019022972.htm |
| [30] |
李亮, 刘杭, 张桂池, 等. 不同低温需求量砂梨休眠相关基因PpMADS13的特征及表达模式[J]. 亚热带植物科学, 2016, 45(1): 42-47. Li L, Liu H, Zhang G C, et al. Characteristics and expression of dormancy-associated MADS-box PpMADS13 in different chilling requirement pear cultivars[J]. Subtropical Plant Science, 2016, 45(1): 42-47 (in Chinese with English abstract). |
| [31] |
Yamane H, Ooka T, Jotatsu H, et al. Expressional regulation of PpDAM5 and PpDAM6, peach(Prunus persica)dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment[J]. Journal of Experimental Botany, 2011, 62(10): 3481-3488. DOI:10.1093/jxb/err028 |
| [32] |
孙艳飞, 闫顺杰, 李勇. 桃树休眠相关基因(DAM)研究进展[J]. 河北果树, 2013(6): 1-2. Sun Y F, Yan S J, Li Y. Research progress of peach dormancy-related gene DAM[J]. Hebei Fruits, 2013(6): 1-2 (in Chinese with English abstract). |
| [33] |
Sasaki R, Yamane H, Ooka T, et al. Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot[J]. Plant Physiology, 2011, 157(1): 485-497. |
| [34] |
Zhao K, Zhou Y Z, Ahmad S, et al. Comprehensive cloning of Prunus mume dormancy associated MADS-box genes and their response in flower bud development and dormancy[J]. Frontiers in Plant Science, 2018, 9: 17. DOI:10.3389/fpls.2018.00017 |
| [35] |
Porto D D, da Silveira Falavigna V, Arenhart R A, et al. Structural genomics and transcriptional characterization of the Dormancy-Associated MADS-box genes during bud dormancy progression in apple[J]. Tree Genetics & Genomes, 2016, 12(3): 46. |
| [36] |
Kumar G, Arya P, Gupta K, et al. Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple(Malus×domestica)[J]. Scientific Reports, 2016, 6: 20695. DOI:10.1038/srep20695 |
| [37] |
Wu R M, Wang T C, Warren B A W, et al. Kiwifruit SVP2 gene prevents premature budbreak during dormancy[J]. Journal of Experimental Botany, 2017, 68(5): 1071-1082. DOI:10.1093/jxb/erx014 |
| [38] |
Wu R M, Wang T C, McGie T, et al. Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time[J]. Journal of Experimental Botany, 2014, 65(17): 4985-4995. DOI:10.1093/jxb/eru264 |
| [39] |
Balogh E, Halász J, Soltész A, et al. Identification, structural and functional characterization of dormancy regulator genes in apricot(Prunus armeniaca L.)[J]. Frontiers in Plant Science, 2019, 10: 402. DOI:10.3389/fpls.2019.00402 |
| [40] |
Diaz-Riquelme J, Lijavetzky D, Martinez-Zapater J M, et al. Genome-wide analysis of MIKCC-type MADS box genes in grapevine[J]. Plant Physiology, 2009, 149(1): 354-369. |
| [41] |
Stockinger E J, Gilmour S J, Thomashow M F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit[J]. Proc Natl Acad Sci USA, 1997, 94(3): 1035-1040. DOI:10.1073/pnas.94.3.1035 |
| [42] |
Prassinos C, Han K H. Characterization of a StMADS11 family gene from sweet cherry(Prunus avium)[C]//American Society of Plant Biologists Annual Meeting. Boston, MA, USA, 2006.
|
| [43] |
Hsu C Y, Adams J P, Kim H, et al. FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar[J]. Proc Natl Acad Sci USA, 2011, 108(26): 10756-10761. DOI:10.1073/pnas.1104713108 |
| [44] |
罗义维.桑树休眠相关MADS-box基因的功能研究[D].重庆: 西南大学, 2018. Luo Y W. Characterization of dormancy related MADS-box genes in mulberry plants[D]. Chongqing: Southwest University, 2018(in Chinese with English abstract). http://d.wanfangdata.com.cn/thesis/D01526998 |
| [45] |
杨堃, 张朝红, 李树秀, 等. 葡萄SVP类MADS-box基因的克隆及表达分析[J]. 西北林学院学报, 2012, 27(4): 117-123. Yang K, Zhang C H, Li S X, et al. Cloning and expression analysis of grape SVP-like MADS-box gene[J]. Journal of Northwest Forestry University, 2012, 27(4): 117-123 (in Chinese with English abstract). |
| [46] |
Busov V B. Plant development:dual roles of poplar SVL in vegetative bud dormancy[J]. Current Biology, 2019, 29(2): R68-R70. DOI:10.1016/j.cub.2018.11.061 |
| [47] |
高真真, 王东岭, 付喜玲, 等. 油桃开花时间调控基因在花芽休眠过程中的表达分析[J]. 园艺学报, 2016, 43(12): 2347-2358. Gao Z Z, Wang D L, Fu X L, et al. Expression analysis of flowering time regulating genes from nectarine flower buds in the process of dormancy[J]. Acta Horticulturae Sinica, 2016, 43(12): 2347-2358 (in Chinese with English abstract). |
| [48] |
Saito T, Bai S L, Imai T, et al. Histone modification and signalling cascade of the dormancy-associated MADS-box gene, PpMADS13-1, in Japanese pear(Pyrus pyrifolia)during endodormancy[J]. Plant, Cell & Environment, 2015, 38(6): 1157-1166. |
| [49] |
Wu R M, Wang T C, Warren B A W, et al. Kiwifruit SVP2 controls developmental and drought-stress pathways[J]. Plant Molecular Biology, 2018, 96(3): 233-244. DOI:10.1007/s11103-017-0688-3 |
| [50] |
Tuan P A, Bai S L, Saito T, et al. Dormancy-associated MADS-box(DAM)and the abscisic acid pathway regulate pear endodormancy through a feedback mechanism[J]. Plant and Cell Physiology, 2017, 58(8): 1378-1390. DOI:10.1093/pcp/pcx074 |
| [51] |
Yamane H, Wada M, Honda C, et al. Overexpression of Prunus DAM6 inhibits growth, represses bud break competency of dormant buds and delays bud outgrowth in apple plants[J]. PLoS One, 2019, 14(4): e0214788. DOI:10.1371/journal.pone.0214788 |
| [52] |
王世祥, 左希亚, 邢利博, 等. 苹果成花抑制蛋白SVP基因的克隆、表达及启动子活性分析[J]. 园艺学报, 2019, 46(8): 1445-1457. Wang S X, Zuo X Y, Xing L B, et al. Cloning, expression pattern and promoter activity analysis of flowering regulatory gene SVP in apple(Malus×domestica)[J]. Acta Horticulturae Sinica, 2019, 46(8): 1445-1457 (in Chinese with English abstract). |
| [53] |
赵倩.草莓SVP同源基因的克隆与表达及其功能分析[D].沈阳: 沈阳农业大学, 2016. Zhao Q. Isolation, expression and function analysis of SVP-like genes in strawberry[D]. Shenyang: Shenyang Agricultural University, 2016(in Chinese with English abstract). http://cdmd.cnki.com.cn/Article/CDMD-10157-1016144964.htm |
| [54] |
Andrés F, Porri A, Torti S, et al. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition[J]. Proc Natl Acad Sci USA, 2014, 111(26): E2760-E2769. DOI:10.1073/pnas.1409567111 |
| [55] |
Wisniewski M, Norelli J, Artlip T. Overexpression of a peach CBF gene in apple:a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants[J]. Frontiers in Plant Science, 2015, 6: 85. |
| [56] |
Zhao K, Zhou Y Z, Ahmad S, et al. PmCBFs synthetically affect PmDAM6 by alternative promoter binding and protein complexes towards the dormancy of bud for Prunus mume[J]. Scientific Reports, 2018, 8: 4527. DOI:10.1038/s41598-018-22537-w |
| [57] |
Yang Q S, Niu Q F, Li J Z, et al. PpHB22, a member of HD-Zip proteins, activates PpDAM1 to regulate bud dormancy transition in 'Suli' pear(Pyrus pyrifolia White Pear Group)[J]. Plant Physiology and Biochemistry, 2018, 127: 355-365. DOI:10.1016/j.plaphy.2018.04.002 |
| [58] |
Wang Q J, Xu G X, Zhao X H, et al. Transcription factor TCP20 regulates peach bud endodormancy by inhibiting PpDAM5/PpDAM6 and interacting with ABF2[J]. Journal of Experimental Botany, 2020, 71(4): 1585-1597. DOI:10.1093/jxb/erz516 |
| [59] |
de la Fuente L, Conesa A, Lloret A, et al. Genome-wide changes in histone H3 lysine 27 trimethylation associated with bud dormancy release in peach[J]. Tree Genetics & Genomes, 2015, 11(3): 45. |
| [60] |
Hiratsu K, Matsui K, Koyama T, et al. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis[J]. The Plant Journal, 2003, 34(5): 733-739. DOI:10.1046/j.1365-313X.2003.01759.x |
| [61] |
Jiménez S, Lawton-Rauh A L, Reighard G L, et al. Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach[J]. BMC Plant Biology, 2009, 9(1): 81. |
| [62] |
Kagale S, Rozwadowski K. EAR motif-mediated transcriptional repression in plants[J]. Epigenetics, 2011, 6(2): 141-146. |
| [63] |
Wu R M, Wang T C, Richardson A C, et al. Histone modification and activation by SOC1-like and drought stress-related transcription factors may regulate AcSVP2 expression during kiwifruit winter dormancy[J]. Plant Science, 2019, 281: 242-250. DOI:10.1016/j.plantsci.2018.12.001 |
| [64] |
Yamane H, Tao R, Ooka T, et al. Comparative analyses of dormancy-associated MADS-box genes, PpDAM5 and PpDAM6, in low-and high-chill peaches(Prunus persica L.)[J]. Journal of the Japanese Society for Horticultural Science, 2011, 80(3): 276-283. DOI:10.2503/jjshs1.80.276 |
| [65] |
da Silveira Falavigna V, Guitton B, Costes E, et al. I want to(bud)break free:the potential role of DAM and SVP-like genes in regulating dormancy cycle in temperate fruit trees[J]. Frontiers in Plant Science, 2019, 9: 1990. DOI:10.3389/fpls.2018.01990 |
| [66] |
牛歆雨, 德庆措姆, 朗杰, 等. 葡萄ARP/DRM基因家族克隆及其在休眠解除过程中的表达[J]. 分子植物育种, 2018, 16(18): 5897-5903. Niu X Y, Deqing C M, Lang J, et al. Cloning of ARP/DRM gene family from grape and its expression during the process of dormancy-release[J]. Molecular Plant Breeding, 2018, 16(18): 5897-5903 (in Chinese with English abstract). |
| [67] |
Ito A, Tuan P A, Saito T, et al. Changes in phytohormone content and associated gene expression throughout the stages of pear(Pyrus pyrifolia Nakai)dormancy[J]. Tree Physiology, 2019. DOI:10.1093/treephys/tpz101 |
| [68] |
Liu J Y, Sherif S M. Hormonal orchestration of bud dormancy cycle in deciduous woody perennials[J]. Frontiers in Plant Science, 2019, 10: 1136. DOI:10.3389/fpls.2019.01136 |
| [69] |
Liu X, Sun Z C, Dong W, et al. Expansion and functional divergence of the SHORT VEGETATIVE PHASE(SVP)genes in eudicots[J]. Genome Biology and Evolution, 2018, 10(11): 3026-3037. DOI:10.1093/gbe/evy235 |
| [70] |
吴璇, 吴少华. 木本植物花芽休眠中激素调节的分子机制研究进展[J]. 武夷学院学报, 2015, 34(3): 53-60. Wu X, Wu S H. Progress of molecular mechanism on hormone regulation of flower bud dormancy in woody plants[J]. Journal of Wuyi University, 2015, 34(3): 53-60 (in Chinese with English abstract). |
| [71] |
Morris C F, Anderberg R J, Goldmark P J, et al. Molecular cloning and expression of abscisic acid-responsive genes in embryos of dormant wheat seeds[J]. Plant Physiology, 1991, 95(3): 814-821. |
| [72] |
Rantanen M, Palonen P. Hot water treatment released endodormancy but reduced number of flowers in potted red raspberry plants[J]. HortScience, 2010, 45(6): 894-898. DOI:10.21273/HORTSCI.45.6.894 |
| [73] |
员盎然.黑穗醋栗(Ribes nigurm L.)二次萌发芽酚类物质鉴定及代谢特征研究[D].哈尔滨: 东北农业大学, 2019. Yuan A R. Identification and metabolic characteristics of phenolic compounds in secondary germination of black currant(Ribes nigurm L.)[D]. Harbin: Northeast Agricultural University, 2019(in Chinese with English abstract). |
| [74] |
Li Y S, Zhou Y Z, Yang W R, et al. Isolation and functional characterization of SVP-like genes in Prunus mume[J]. Scientia Horticulturae, 2017, 215: 91-101. DOI:10.1016/j.scienta.2016.12.013 |




