文章信息
- 谷涛, 李永丰, 周潮洋, 张迹, 闫新, 李顺鹏
- GU Tao, LI Yongfeng, ZHOU Chaoyang, ZHANG Ji, YAN Xin, LI Shunpeng
- 取代脲类除草剂降解菌鞘氨醇杆菌(Sphingobium sp.)Pu21的分离和鉴定与降解特性
- Isolation, identification and biodegradation characteristics of phenylurea herbicide-degrading bacterium Sphingobium sp.Pu21
- 南京农业大学学报, 2017, 40(4): 655-663
- Journal of Nanjing Agricultural University, 2017, 40(4): 655-663.
- http://dx.doi.org/10.7685/jnau.201609042
-
文章历史
- 收稿日期: 2016-09-28
2. 南京农业大学农业部农业环境微生物工程重点开放实验室, 江苏 南京 210095;
3. 淮阴师范学院江苏省环洪泽湖生态农业生物技术重点实验室, 江苏 淮安 223300
2. Key Laboratory of Microbiological Engineering Agricultural Environment, Ministry of Agriculture, Nanjing Agricultural University, Nanjing 210095, China;
3. Jiangsu Key Laboratory for Eco-Agricultural Biotechnology around Hongze Lake, Huaiyin Normal University, Huai'an 223300, China
取代脲类除草剂包括异丙隆、敌草隆、绿麦隆、利谷隆等。根据脲分子中氨基侧链上取代基的不同分为两类:一类是N, N-二甲基取代脲类除草剂(如异丙隆、敌草隆和绿麦隆等); 另一类是N-甲氧基-N-甲基取代脲类除草剂(利谷隆和绿谷隆等), 主要用来防除1年生禾本科杂草和阔叶杂草[1-2], 因其高效、广谱而被广泛应用于农业生产中。取代脲类除草剂具有较高的水溶性和较低的土壤吸附性, 容易迁移运动。多种该类除草剂及其代谢产物污染物常在地下水[3]、河流和小溪[4]、湖泊[5]以及海水[6]中检测到。据研究表明, 该类除草剂及其代谢产物残留对微生物、动植物及人类的健康造成了极大的危害[7-9]。释放到环境中的取代脲类除草剂可通过多种途径移除, 包括光解、水解、羟基化、氧化和微生物降解等[10-13]。由于此类除草剂化学性质稳定, 非生物降解速度慢, 因此微生物降解是去除环境中取代脲类除草剂的最主要途径。目前, 研究人员已从环境中分离出大量取代脲类除草剂的降解菌, 主要包括鞘氨醇杆菌属(Sphingobium)[14]、鞘氨醇单胞菌属(Sphingomonas)[15-18]、假单胞菌属(Pseudomonas)[19-20]、节杆菌属(Arthrobacter)[21]、芽孢杆菌属(Bacillus)[22]、贪噬菌属(Variovorax)[23]、微球菌属(Micrococcus)[24]等, 但其中的高效、广谱降解菌株并不多, 特别是兼降2类取代脲类除草剂的菌株更为稀少。因此分离筛选更优良的降解菌株, 具有重要的现实意义。本试验分离到1株能同时降解异丙隆和利谷隆的高效降解菌, 并对其降解特性和降解途径进行了初步研究, 为取代脲类除草剂污染的生物修复提供了优良的菌株资源。
1 材料与方法 1.1 供试材料 1.1.1 主要仪器MS/MS串联质谱仪(Finnigan TSQ Quantum Ultra AM, Thermal, USA); SPD-20A型高效液相色谱仪(日本岛津公司); 紫外-可见分光光度计(岛津UV-2401PCP/450);EDC810 PCR仪(北京东胜创新生物科技有限公司); DYCP-31DN型电泳仪(北京市六一仪器厂); DNA提取试剂盒(北京全式金生物技术有限公司); AxyPrep DNA凝胶回收试剂盒(爱思进生物技术有限公司)。
1.1.2 试剂异丙隆原药(纯度>97%)、绿麦隆原药(纯度>97%)、敌草隆原药(纯度>98%)、利谷隆原药(纯度>97%)和伏草隆原药(纯度>97%)由江苏快达农化股份有限公司友情赠送。甲氧隆、灭草隆、非草隆、绿谷隆购于百灵威科技有限公司。所有原药和标样用甲醇溶解配置成10 g·L-1储存液备用。高效液相色谱(HPLC)分析用的甲醇为色谱纯试剂, 天津科密欧化学试剂有限公司生产。二氯甲烷等其他化学试剂均为普通的分析纯试剂。
1.1.3 土壤样品样品采集于长期生产取代脲类除草剂的江苏某化工厂, 为污泥样品, 取回后于-20 ℃保存备用。
1.1.4 培养基LB培养基:蛋白胨10.0 g·L-1, 酵母膏5.0 g·L-1, NaCl 10.0 g·L-1, pH7.0;基础盐培养基(MSM):NaCl 1.0 g·L-1, NH4NO3 1.0 g·L-1, K2HPO4 1.5 g·L-1, KH2PO4 0.5 g·L-1, MgSO4·7H2O 0.2 g·L-1, pH7.0。
1.2 降解菌株的富集与分离采自农药厂的2 g污泥样品置于异丙隆质量浓度为20 mg·L-1的100 mL基础盐培养基中, 于30 ℃、150 r·min-1富集培养。每隔5 d以10%(体积分数)的接种量转接到新鲜培养基中, 连续富集3次。培养液经紫外-可见分光光度计测定降解效果, 有效果的富集液进行梯度稀释后, 在含100 mg·L-1异丙隆的基础盐培养基固体平板上涂布0.5 mL, 于30 ℃培养箱培养, 待平板上出现单菌落后, 挑取单菌落分别转接至含20 mg·L-1异丙隆和利谷隆的MSM液体培养基中, 于30 ℃、150 r·min-1摇床培养7 d, HPLC检测其降解异丙隆和利谷隆的效果。
1.3 菌株的鉴定 1.3.1 形态及生理生化指标鉴定菌株形态及生理生化鉴定方法参见文献[25]。
1.3.2 菌株16S rRNA基因PCR扩增及序列的测定以提取的菌株总DNA为模板, 进行16S rRNA基因的PCR扩增, 正向引物:5′-AGAGTTTGATCCTGGCTCAG-3′(Escherichia coli bases 8 to 27);反向引物:5′-TACCTTGTTACGACT T-3′(Escherichia coli bases 1 507 to 1 492)。PCR反应体系(50 μL):10×Taq缓冲液5 μL, dNTP(25 mmol·L-1)4 μL, MgCl2(25 mmol·L-1)4 μL, 菌体DNA(约50 ng·μL-1)1 μL, 引物(25 μmol·L-1)各1 μL, Taq DNA聚合酶(5 U·L-1)0.5 μL, ddH2O 33.5 μL。PCR反应条件为:95 ℃ 5 min; 94 ℃ 30 s, 55 ℃ 30 s, 72 ℃ 1.5 min, 30个循环; 72 ℃延伸10 min。琼脂糖凝胶电泳检测扩增产物的大小(1.5 kb左右), 并用凝胶回收试剂盒回收PCR产物, T/A克隆后进行测序, 将测定的16S rRNA基因序列与EzTaxon-e(http://www.ezbiocloud.net/eztax)数据库[26]中相关标准菌株和本实验室分离相关菌株Sphingobium spp.YBL1、YBL2、YBL3和Sphingomonas sp.Y57的16S rRNA基因序列进行同源性比对分析。应用MEGA 5.0软件, 采用CLUSTAL X将所有序列比对排序后, 用软件中的Kimura2-parameter Distance模型计算进化距离, Neighbor-Joining法构建系统发育树, 5 000次随机抽样, 计算自引导值(Bootstrap)以评估发生树的置信度。
1.4 环境因素对菌株降解性能的影响 1.4.1 种子液的制备挑取在LB培养基固体平板上的单菌落, 接种到100 mL LB液体培养基中, 30 ℃、150 r·min-1摇床振荡培养约48 h, 离心收集菌体, 用MSM洗涤2次后重悬浮, 并调整菌液浓度为D600至2.5, 即为种子液。
1.4.2 温度对菌株降解异丙隆和利谷隆的影响在MSM培养基中加入30 mg·L-1异丙隆或利谷隆, 以2%(体积分数)接种量接入降解菌株种子液, 分别在15、20、25、30、35和40 ℃下, 于150 r·min-1摇床中培养, 异丙隆于24 h取样, 利谷隆4 d后取样, 检测农药降解情况。所有试验设3个平行。
1.4.3 初始pH值对降解的影响调节MSM培养基的pH分别为4、5、6、7、8、9和10, 然后分别加入30 mg·L-1异丙隆或利谷隆, 以2%接种量接入种子液, 置于30 ℃、150 r·min-1摇床中培养, 异丙隆于24 h取样, 利谷隆于4 d后取样, 检测农药降解情况。所有试验设3个平行。
1.4.4 外加碳、氮源对降解影响在含有30 mg·L-1异丙隆或利谷隆的MSM培养基中, 分别添加已灭过菌的葡萄糖、麦芽糖、蛋白胨和酵母粉至含量为5 g·L-1, 以2%接种量接入种子液, 置于30 ℃、150 r·min-1摇床中培养。异丙隆于24 h取样, 利谷隆于4 d后取样, 检测样品中农药的降解情况。所有试验设3个平行。以不加外源碳、氮的处理为对照。
1.5 Pu21的底物降解谱在MSM培养基中分别添加20 mg·L-1异丙隆、敌草隆、伏草隆、绿麦隆、甲氧隆、灭草隆、非草隆、利谷隆和绿谷隆作为唯一碳源, 按2%接种量接入降解菌株Pu21的种子液, 30 ℃、150 r·min-1摇床培养, 于7 d后取样, 测定各样品的降解情况, 各样品均设不接菌的空白对照。
1.6 Pu21以异丙隆和利谷隆为唯一碳源的生长降解及降解途径的鉴定在100 mL MSM培养基中以2%接种量接种种子液, 并向其中加入异丙隆或利谷隆的母液, 使其终质量浓度为30 mg·L-1。将菌悬液置于恒温摇床中, 30 ℃、150 r·min-1振荡培养, 定时取样(添加异丙隆处理每6 h取样1次, 利谷隆每1 d取样1次), 设接种灭活的Pu21为对照。样品处理后菌体生长量采用比浊法测定, 即检测600 nm的吸光值。异丙隆和利谷隆的含量及其代谢产物分别用液相色谱和质谱检测。
1.7 取代脲类除草剂的测定取代脲类除草剂的降解及中间代谢物的检测通过紫外-可见分光光度计、高效液相色谱仪(HPLC)和质谱仪测定。样品提取:在待测样品中加入等体积的二氯甲烷, 充分振荡, 静置分层后弃去上层水相, 有机相经无水硫酸钠脱水后, 取1 mL有机相置于1.5 mL离心管中, 吹干后加入0.5 mL甲醇(色谱纯)溶解, 经孔径0.45 μm的有机相滤器过滤后, 待测。分光光度计扫描:扫描波长为200~350 nm, 检测底物的特征吸收峰是否变化, 以此来确定菌株的降解能力。液相色谱分析:色谱柱为Kromasil 100-5C18(内径:4.6 mm; 长:25 cm), 流动相为:甲醇与水(体积比为80 : 20), 流速为0.8 mL·min-1, 检测波长分别为230和250 nm, 进样量为20 μL。根据各个物质对应的峰面积, 参照已知浓度对照标准物峰面积计算每个样品中底物及产物的浓度。质谱分析:质谱为美国Finnigan MS/MS串联质谱仪, 配有ESI离子源。在质谱检测过程中, 代谢产物经阳性电极离子源电喷射被离子化并带1个正电荷, 通过一级质谱分离并确定离子分子质量[M+H]+。代谢产物的特征离子峰再次通过二级质谱分析。
2 结果与分析 2.1 菌株的分离与鉴定 2.1.1 菌株的生理生化特性经过分离纯化得到1株异丙隆降解菌Pu21, 该菌株在LB平板上生长3 d后, 形成淡黄色、圆形、不透明、边缘整齐、表面隆起、湿润光滑的菌落。菌株Pu21为革兰氏阴性细菌, 不运动, 不产芽孢, 严格好氧。氧化酶、过氧化氢酶和硝基还原酶阳性, 脲酶和淀粉酶阴性, 不能水解明胶和七叶苷。利用葡萄糖、甘露醇、木糖、阿拉伯糖生长时产酸。菌株对低于100 mg·L-1的氨苄青霉素、羧苄青霉素和链霉素具有抗性, 而对庆大霉素、卡那霉素、壮观霉素、氯霉素、四环素和红霉素等抗生素敏感。
2.1.2 菌株16S rRNA基因序列分析以菌株Pu21的总DNA为模板, 用16S rRNA通用引物进行PCR扩增, 得到长度为1.5 kb的扩增产物, 测序后在GenBank登录, 序列号为KM386662。与其他菌株16S rRNA基因序列同源性比较结果(图 1)显示:与菌株Pu21的16S rRNA基因同源性最高的是Sphingobium属, 其中与Sphingobium sp.YBL3[14]相似性为99%, 与Sphingobium fuliginis TKPT的序列相似性为98%。结合细菌形态、生理生化、16S rRNA基因系统发育分析将菌株Pu21初步鉴定为鞘氨醇杆菌属(Sphingobium sp.)。

|
图 1 菌株Pu21基于16S rRNA基因序列同源性的系统发育树分析 Figure 1 Phylogenetic analysis of strain Pu21 and related species by the neighbor joining approach |
从图 2可知:菌株Pu21能以异丙隆为唯一碳源生长, 36 h时降解率达99%以上。除了异丙隆, Pu21还能以利谷隆为唯一碳源生长, 但降解利用利谷隆速度较慢, 降解30 mg·L-1的利谷隆需要7 d。伴随着利谷隆的降解, 菌株Pu21的生物量缓慢提高, 最后趋于稳定。

|
图 2 菌株Pu21以异丙隆(A)和利谷隆(B)为唯一碳源生长和降解情况 Figure 2 Utilization of isoproturon(A)and linuron(B)as sole source of carbon for growth by strain Pu21 |
35 ℃时菌株Pu21对异丙隆和利谷隆的降解最快; 25~35 ℃范围内温度变化对菌株Pu21降解异丙隆和利谷隆影响较小; 当温度低于20 ℃或高于40 ℃时降解效果显著下降, 异丙隆和利谷隆的降解率只有20%~30%左右(图 3)。

|
图 3 温度对菌株降解异丙隆和利谷隆的影响 Figure 3 Effect of temperature on isoproturon and linuron degradation by strain Pu21 |
试验结果(图 4)显示:菌株在pH7.0的培养基中降解异丙隆和利谷隆速率最快; 菌株在pH为6~8时皆能较快降解异丙隆和利谷隆; 当培养基的pH值为5和9时, 菌株对农药的降解速率明显下降; 当pH值小于5或者是大于9时, 几乎不能降解异丙隆和利谷隆。
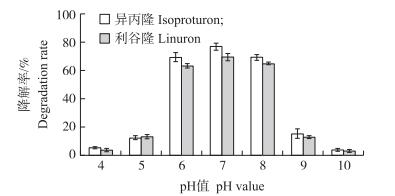
|
图 4 初始pH值对菌株降解异丙隆和利谷隆的影响 Figure 4 Effect of initial pH on isoproturon and linuron degradation by strain Pu21 |
加入葡萄糖和麦芽糖时, 对菌株降解异丙隆基本没有影响, 但利谷隆降解显著提高; 在加入蛋白胨和酵母粉的处理中, 异丙隆和利谷隆的降解率都有显著的提高, 尤其是对利谷隆的降解(图 5)。

|
图 5 不同碳、氮源对菌株降解异丙隆和利谷隆的影响 Figure 5 Effect of carbon and nitrogen source on isoproturon and linuron degradation by stain Pu21 |
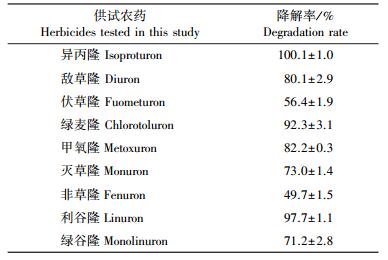
为了研究菌株Pu21的底物谱, 选择了9种常见的取代脲类除草剂, 结果(表 1)显示:菌株Pu21可以降解所有的供试农药, 7 d后加入的异丙隆被完全降解, 利谷隆被降解了97.7%, 但对伏草隆和非草隆的降解效果较差(伏草隆被降解56.4%, 非草隆被降解49.7%)。所有不接菌的对照, 都没检测到农药的降解。
菌株Pu21降解异丙隆过程中, HPLC检测到2个代谢产物, 保留时间分别为5.7和6.2 min(图 6-A)。质谱检测发现有4个比较明显的分子离子峰, 质荷比m/z(M+H)分别为207.09、193.07、179.06和136.11(图 6-A)。用二级质谱分析这4个分子离子峰, 除了207.09为底物异丙隆外, 其他产物特征碎片离子峰分别见图 6-B、C、D。根据这些产物对应的二级质谱特征碎片峰, 这3个物质分别被鉴定为1-(4-异丙基苯基)-3-甲基脲(MDIPU)、1-对异丙基苯基脲(DDIPU)和对异丙基苯胺(4IA)。推测该菌株降解异丙隆的途径与已报道的异丙隆降解菌株Sphingomonas sp.SRS2和Sphingobium sp.YBL2的途径相同[27-28], 即异丙隆N-基团上先脱甲基生成MDIPU, 然后再脱甲基生成DDIPU, 接着断开脲侧链形成4IA。

|
图 6 菌株Pu21降解异丙隆的液相色谱和质谱分析 Figure 6 HPLC and MS/MS analysis of isoproturon transformation by strain Pu21 A为降解产物的HPLC图和一级质谱图; B、C和D分别是m/z为193.07、179.06和136.11物质的二级质谱图。 The metabolites were detected using HPLC and standard MS(A). The m/z=193.07[M+H]+(B), m/z=179.06[M+H]+(C)and m/z=136.11[M+H]+(D)characteristic ion peaks were subjected to second-order MS analysis and were characterized as MDIPU, DDIPU and 4IA, respectively. |
菌株Pu21降解利谷隆时, HPLC分析只检测到1个代谢产物, 保留时间为6.414 min(图 7-A)。将有明显代谢产物积累的样品进行MS/MS分析, 在一级质谱中形成了2个比较突出的分子离子峰, 质荷比m/z(M+H)分别为248.91和161.95。将这2种物质对应的分子离子峰进一步用二级质谱分析, 得到的特征碎片峰见图 8-B和C, 经鉴定这2种物质分别为利谷隆和3, 4-二氯苯胺(3, 4-DCA)。结合已经报道利谷隆的代谢途径[23], 推测Pu21降解利谷隆是直接断脲桥生成3, 4-DCA。
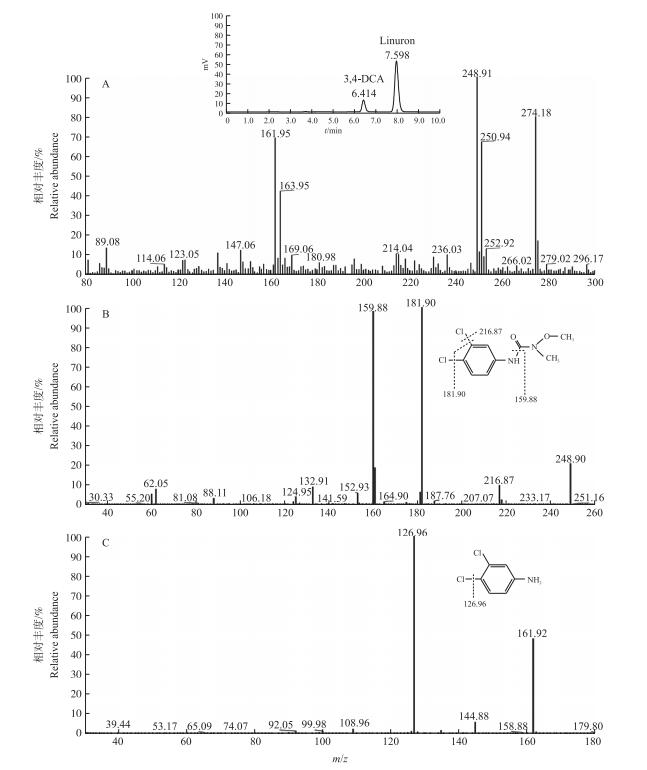
|
图 7 菌株Pu21降解利谷隆的液相色谱和质谱分析 Figure 7 HPLC and MS/MS analysis of linuron transformation by strain Pu21 A为降解产物的HPLC图和一级质谱图; B和C分别是m/z为248.91和161.95物质的二级质谱图。 The metabolites were detected using HPLC and standard MS(A). The m/z=248.91[M+H]+(B)and m/z=161.92[M+H]+(C)characteristic ion peaks were subjected to second-order MS analysis and were characterized as linuron and 3, 4-DCA, respectively. |
目前, 国内外已报到的可以降解取代脲类除草剂的微生物很多。早在1969年, Wallnöfer等[29]就分离到1株能够降解绿谷隆和溴谷隆的降解菌Bacillis sphaericus ATCC12123, 但该菌不能降解N, N-二甲基取代脲类除草剂。此后, 陆续分离到许多降解菌株, 主要可以分为3类:第1类, 只能降解N, N-二甲基取代脲类除草剂的降解菌株, 如Sphingomonas sp.SRS2[15]; 第2类, 只能降解N-甲氧基-N-甲基取代脲类除草剂的降解菌株, 如Variovorax sp.RA8[30]; 第3类, 能降解2类取代脲类除草剂的菌株。Arthrobacter globiformis D47是1株高效的取代脲类除草剂的降解菌, 能够降解大多数的取代脲类除草剂, 降解的方式是直接断脲桥生成相应的苯胺衍生物。该菌降解利谷隆的速度最快, 其次是敌草隆, 降解异丙隆的速度很慢, 只有在加入额外的碳、氮源后, 才能提高降解异丙隆的速度[31]。Variovorax sp.SRS16能较快降解利谷隆, 在加入其他底物时, 该菌还能够降解低浓度的敌草隆[32]。阳性菌Micrococcus sp.PS-1能够降解较高浓度的敌草隆(250 mg·L-1), 降解的途径是脱甲基后断脲桥生成3, 4-二氯苯胺。该菌还能降解N-甲氧基-N-甲基类取代脲类除草剂, 如利谷隆和绿谷隆[24], 但没有研究该菌株降解利谷隆和绿谷隆的代谢途径。
菌株Sphingobium spp.YBL1、YBL2和YBL3及Sphingomonas sp.Y57均能高效降解N, N-二甲基类取代脲类除草剂, 其中YBL1还能够非常缓慢地降解利谷隆, 30 d能将10 mg·L-1利谷隆降解60%[14, 17]。菌株Pu21与上述几株菌存在明显差别:YBL1、YBL2和YBL3对100 mg·L-1氨苄青霉素敏感, Pu21对氨苄青霉素不敏感。另外, 菌株Pu21在降解性能方面与上述几株菌显著不同, Pu21能在7 d之内将30 mg·L-1利谷隆完全降解, 速度远超YBL1[14]。通过HPLC和MS/MS鉴定异丙隆和利谷隆降解过程中的代谢产物, 推断了菌株Pu21降解异丙隆和利谷隆的代谢途径, 虽然这2种降解途径已有报道, 但是以这种方式降解两类除草剂的降解菌株尚未见报道。
国内外关于这方面的工作, 除了分离降解菌株以外, 在降解酶和基因方面也进行了研究。Engelhardt等[33]从菌株Bacillus sphaericus ATCC12123中纯化出相对分子质量为75×103的芳基酰基酰胺酶, 该酶能切断利谷隆等N-甲氧基-N-甲基苯基脲类除草剂的脲桥, 但是不能作用于异丙隆、敌草隆等N, N-二甲类苯基脲除草剂的脲桥, 也未克隆到相关的基因; 克隆自Arthrobacter globiformis D47和Mycobacterium brisbanense JK1中的水解酶基因puhA和puhB具有较高的同源性, 编码的酶可以切断大多数苯基脲类除草剂的脲桥, 但对N, N-二甲基取代脲类除草剂的催化活性较低[34]。Bers等[35]从利谷隆的降解菌株Variovorax sp.WDL1中克隆到1个水解酶基因hylA, 该基因编码的蛋白能够将利谷隆的脲桥断裂生成3, 4-DCA。从YBL2中克隆到了负责从N, N-二甲基取代脲类除草剂上脱甲基的基因pdmAB和负责DDIPU断脲桥的基因ddhA, 并发现这些基因在菌株YBL1、YBL2、YBL3和Pu21[36-37]中高度保守, 但菌株Pu21中利谷隆的降解基因有待进一步研究。
| [1] | Greulich K, Hoque E, Pflugmacher S. Uptake, metabolism, and effects on detoxication enzymes of isoproturon in spawn and tadpoles of amphibians[J]. Ecotoxicology and Environmental Safety, 2002, 52(3): 256–266. DOI: 10.1006/eesa.2002.2182 |
| [2] | Chhokar R S, Singh S, Sharma R K. Herbicides for control of isoproturon-resistant Littleseed Canarygrass(Phalaris minor)in wheat[J]. Crop Protection, 2008, 27(3): 719–726. |
| [3] | Field J A, Reed R L, Sawyer T E, et al. Diuron and its metabolites in surface water and ground water by solid phase extraction and in-vial elution[J]. Journal of Agricultural and Food Chemistry, 1997, 45(10): 3897–3902. DOI: 10.1021/jf970196v |
| [4] | Spliid N H, Køppen B. Occurrence of pesticides in Danish shallow ground water[J]. Chemosphere, 1998, 37(7): 1307–1316. DOI: 10.1016/S0045-6535(98)00128-3 |
| [5] | Thurman E M, Bastian K C, Mollhagen T. Occurrence of cotton herbicides and insecticides in playa lakes of the High Plains of West Texas[J]. Science of the Total Environment, 2000, 248(2): 189–200. |
| [6] | Gerecke A C, Tixier C, Bartels T, et al. Determination of phenylurea herbicides in natural waters at concentrations below 1 ng·L-1 using solid-phase extraction, derivatization, and solid-phase microextraction-gas chromatography-mass spectrometry[J]. Journal of Chromatography A, 2001, 930(1): 9–19. |
| [7] | Cycon M, Piotrowska-Seget Z, Kozdroj J. Linuron effects on microbiological characteristics of sandy soils as determined in a pot study[J]. Annals of Microbiology, 2010, 60: 439–449. DOI: 10.1007/s13213-010-0061-0 |
| [8] | Lopez-Doval J C, Ricart M, Guasch H, et al. Does grazing pressure modify diuron toxicity in a biofilm community?[J]. Archives of Environmental Contamination and Toxicology, 2010, 58: 955–962. DOI: 10.1007/s00244-009-9441-5 |
| [9] | Da-Rocha M S, Nascimento M G, Cardoso A P, et al. Cytotoxicity and regenerative proliferation as the mode of action for diuron-induced urothelial carcinogenesis in the rat[J]. Toxicological Sciences, 2010, 113: 37–44. DOI: 10.1093/toxsci/kfp241 |
| [10] | Katsumata H, Sada M, Nakaoka Y, et al. Photocatalytic degradation of diuron in aqueous solution by platinized TiO2[J]. Journal of Hazardous Materials, 2009, 171: 1081–1087. DOI: 10.1016/j.jhazmat.2009.06.110 |
| [11] | Reddy P A K, Reddy P V L, Sharma V M, et al. Photocatalytic degradation of isoproturon pesticide on C, N and S Doped TiO2[J]. Journal of Water Resource and Protection, 2010, 2(3): 235–244. DOI: 10.4236/jwarp.2010.23027 |
| [12] | Gangwar S K, Rafiquee M Z A. Kinetics of the alkaline hydrolysis of isoproturon in CTAB and NaLS micelles[J]. International Journal of Chemical Kinetics, 2007, 39: 39–45. DOI: 10.1002/(ISSN)1097-4601 |
| [13] | Vicente F, Santos A, Romero A, et al. Kinetic study of diuron oxidation and mineralization by persulphate:effects of temperature, oxidant concentration and iron dosage method[J]. Chemical Engineering Journal, 2011, 170: 127–135. DOI: 10.1016/j.cej.2011.03.042 |
| [14] | Sun J Q, Huang X, Chen Q L, et al. Isolation and characterization of three Sphingobium sp.strains capable of degrading isoproturon and cloning of the catechol 1, 2-dioxygenase gene from these strains[J]. World Journal of Microbiology and Biotechnology, 2009, 25(2): 259–268. DOI: 10.1007/s11274-008-9888-y |
| [15] | Sørensen S R, Aamand J. Biodegradation of the phenylurea herbicide isoproturon and its metabolites in agricultural soils[J]. Biodegradation, 2001, 12(1): 69–77. DOI: 10.1023/A:1011902012131 |
| [16] | Bending G D, Lincoln S D, Sørensen S R, et al. In-field spatial variability in the degradation of the phenyl-urea herbicide isoproturon is the result of interactions between degradative Sphingomonas spp. and soil pH[J]. Applied and Environmental Microbiology, 2003, 69(2): 827–834. DOI: 10.1128/AEM.69.2.827-834.2003 |
| [17] |
孙纪全, 黄星, 何健, 等. 异丙隆降解菌Y57的分离鉴定及其降解特性[J].
中国环境科学, 2006, 26(3): 315–319.
Sun J Q, Huang X, He J, et al. Isolation identification of isoproturon degradation bacterium Y57 and its degradation characteristic[J]. China Environmental Science, 2006, 26(3): 315–319. (in Chinese with English abstract) |
| [18] | Hussain S, Devers-Lamrani M, El A N, et al. Isolation and characterization of an isoproturon mineralizing Sphingomonas sp.strain SH from a French agricultural soil[J]. Biodegradation, 2011, 22(3): 637–650. DOI: 10.1007/s10532-010-9437-x |
| [19] | El-Deeb B A, Soltan S M, Ali A M, et al. Detoxication of the herbicide diuron by Pseudomonas sp.[J]. Folia Microbiologica, 2000, 45(3): 211–216. DOI: 10.1007/BF02908946 |
| [20] | Batisson I, Pesce S, Besse-Hoggan P, et al. Isolation and characterization of diuron-degrading bacteria from lotic surface water[J]. Microbial Ecology, 2007, 54(4): 761–770. DOI: 10.1007/s00248-007-9241-2 |
| [21] | Tixier C, Sancelme M, Ait-Aissa S, et al. Biotransformation of phenylurea herbicides by a soil bacterial strain Arthrobacter sp.N2:structure, ecotoxicity and fate of diuron metabolite with soil fungi[J]. Chemosphere, 2002, 46(4): 519–526. DOI: 10.1016/S0045-6535(01)00193-X |
| [22] | Ngigi A, Getenga Z, Boga H, et al. Biodegradation of phenylurea herbicide diuron by microorganisms from long-term-treated sugarcane-cultivated soils in Kenya[J]. Toxicological and Environmental Chemistry, 2011, 93(8): 1623–1635. DOI: 10.1080/02772248.2011.595718 |
| [23] | Dejonghe W, Berteloot E, Goris J, et al. Synergistic degradation of linuron by a bacterial consortium and isolation of a single linuron-degrading Variovorax strain[J]. Applied and Environmental Microbiology, 2003, 69(3): 1532–1541. DOI: 10.1128/AEM.69.3.1532-1541.2003 |
| [24] | Sharma P, Chopra A, Cameotra S S, et al. Efficient biotransformation of herbicide diuron by bacterial strain Micrococcus sp.PS-1[J]. Biodegradation, 2010, 21(6): 979–987. DOI: 10.1007/s10532-010-9357-9 |
| [25] |
东秀珠, 蔡妙英.
常见细菌系统鉴定手册[M].北京: 科学出版社, 2001.
Dong X Z, Cai M Y. Manual of Common System Determination Bacteriology[M].Beijing: Science Press, 2001. (in Chinese with English abstract) |
| [26] | Kim O S, Cho Y J, Lee K, et al. Introducing EzTaxon-e:a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species[J]. International Journal of Systematic and Evolutionary Microbiology, 2012, 62: 716–721. DOI: 10.1099/ijs.0.038075-0 |
| [27] | Sørensen S R, Ronen Z, Aamand J. Isolation from agricultural soil and characterization of a Sphingomonas sp.able to mineralize the phenylurea herbicide isoproturon[J]. Applied and Environmental Microbiology, 2002, 67(12): 5403–5409. |
| [28] | Zhang J, Hong Q, Li Q, et al. Characterization of isoproturon biodegradation pathway in Sphingobium sp.YBL2[J]. International Biodeterioration and Biodegradation, 2012, 70(5): 8–13. |
| [29] | Wallnöfer P. The decomposition of urea herbicides by Bacillus sphaericus, isolated from soil[J]. Weed Research, 1969, 9(4): 333–339. DOI: 10.1111/wre.1969.9.issue-4 |
| [30] | Satsuma K. Mineralisation of the herbicide linuron by Variovorax sp.strain RA8 isolated from Japanese river sediment using an ecosystem model(microcosm)[J]. Pest Management Science, 2010, 66(8): 847–852. |
| [31] | Cullington J E, Walker A. Rapid biodegradation of diuron and other phenylurea herbicides by a soil bacterium[J]. Soil Biology and Biochemistry, 1999, 31(5): 677–686. DOI: 10.1016/S0038-0717(98)00156-4 |
| [32] | Sørensen S R, Rasmussen J, Jacobsen C S, et al. Elucidating the key member of a linuron-mineralizing bacterial community by PCR and reverse transcription-PCR denaturing gradient gel electrophoresis 16S rRNA gene fingerprinting and cultivation[J]. Applied and Environmental Microbiology, 2005, 71(7): 4144–4148. DOI: 10.1128/AEM.71.7.4144-4148.2005 |
| [33] | Engelhardt G, Wallnöfer P R, Plapp R. Purification and properties of an aryl acylamidase of Bacillus sphaericus, catalyzing the hydrolysis of various phenylamide herbicides and fungicides[J]. Applied Microbiology, 1973, 26(5): 709–718. |
| [34] | Khurana J, Jackson C, Scott C, et al. Characterization of the phenylurea hydrolases A and B:founding members of a novel amidohydrolase subgroup[J]. Biochemical Journal, 2009, 418: 431–441. DOI: 10.1042/BJ20081488 |
| [35] | Bers K, Batisson I, Proost P, et al. HylA, an alternative hydrolase for initiation of catabolism of the phenylurea herbicide linuron in Variovorax sp. strains[J]. Applied and Environmental Microbiology, 2013, 79(17): 5258–5263. DOI: 10.1128/AEM.01478-13 |
| [36] | Gu T, Zhou C, Sørensen S R, et al. The novel bacterial N-demethylase PdmAB is responsible for the initial step of N, N-dimethyl-substituted phenylurea herbicide degradation[J]. Applied and Environmental Microbiology, 2013, 79(24): 7846–7856. DOI: 10.1128/AEM.02478-13 |
| [37] | Yan X, Gu T, Yi Z, et al. Comparative genomic analysis of isoproturon-mineralizing sphingomonads reveals the isoproturon catabolic mechanism[J]. Environmental Microbiology, 2016, 18(12): 4888–4906. DOI: 10.1111/1462-2920.13413 |