文章信息
- 李慧, 顾天滋, 陈昌宇, 黄开茹, 田朔, 赵旭东, 郝德君
- Li Hui, Gu Tianzi, Chen Changyu, Huang Kairu, Tian Shuo, Zhao Xudong, Hao Dejun
- 仁扇舟蛾化学感受蛋白cDNA的克隆、序列分析及时空表达
- cDNA Cloning, Sequence Analysis and Expression Profile of a Chemosensory Protein from the Clostera restitura (Lepidoptera: Notodontidae)
- 林业科学, 2018, 54(4): 67-75.
- Scientia Silvae Sinicae, 2018, 54(4): 67-75.
- DOI: 10.11707/j.1001-7488.20180408
-
文章历史
- 收稿日期:2017-02-27
- 修回日期:2017-12-19
-
作者相关文章
昆虫在长期进化中形成了高度特异、极其灵敏的嗅觉感受系统,借助复杂而精确的嗅觉感受系统,昆虫可以感知环境中的各种化学信息,并将这些外界的化学信息转化为体内的电信号,启动特定的信号传导途径,进而产生觅食、求偶、交配、繁殖、防御、迁徙等行为(王桂荣等,2002;Brito et al., 2016)。嗅觉的识别过程中会产生一系列的生理反应,而这些反应离不开多种蛋白的参与,如气味结合蛋白(odorant binding proteins, OBPs)、化学感受蛋白(chemosensory proteins, CSPs)、气味受体(odor receptors, ODs)、气味降解酶(odor degrading enzymes, ODEs)、离子型受体(ionic receptors, IRs)、感觉神经元膜蛋白(sensory neuron membrane proteins, SNMPs)等(Vogt et al., 1985; 1991; 2009)。每种蛋白都发挥其不可或缺的作用,其中作为环境信息结合载体有2类水溶性蛋白,即气味结合蛋白和化学感受蛋白(Pelosi et al., 2006)。气味结合蛋白的功能是结合运输挥发性的气味分子和性信息素,而化学感受蛋白主要结合并转运非挥发性的气味分子(Steinbrecht et al., 1995)。
自从Mc Kenna(1994)首次从果蝇(Drosophila melanogaster)触角中克隆出CSP Dmelos-d结构以来,研究者们已在鳞翅目(Lepidoptera) (张天涛等,2011;邓培渊等,2015;郭兴国等,2016)、膜翅目(Hymenoptera) (李红亮等,2010)、直翅目(Orthoptera) (Picimbon et al., 2000)、半翅目(Hemiptera) (马双等,2011;赵洁等,2015)、双翅目(Diptera) (Pelletier et al., 2011)、鞘翅目(Coleoptera) (张鑫鑫,2015)、蜚蠊目(Blattaria) (Picimbon et al., 1999)、竹节虫目(Phasmatodea) (Marchese et al., 2000)、脉翅目(Neuroptera) (王思豹等,2015)中均有发现。关于鳞翅目昆虫的化学感受蛋白已有广泛和深入研究,先后在棉铃虫(Helicoverpa armigera)、烟青虫(Heliothis assulta)、亚洲玉米螟(Ostrinia furnacalis)、双委夜蛾(Athetis dissimilis)以及小菜蛾(Plutella xylostella)等种类中发现(张志春等,2009; 张天涛等,2011;Zhang et al., 2015;Sun et al., 2016; Yang et al., 2016)。化学感受蛋白在多种昆虫中都有所分布,表明其在嗅觉识别过程中起着不可缺少的作用,并且这一功能极其普遍。
昆虫的化学感受蛋白由约120个氨基酸组成,分子量大约在13~17 kDa,具有4个半胱氨酸保守位点,含有1个CX6CX17-19CX2C结构域,2个二硫键,相邻的2个半胱氨酸形成二硫键,从而形成稳定的结构(Calvello et al., 2005; Xu et al., 2009; Pelosi et al., 2014)。化学感受蛋白是可溶性球蛋白,二级结构主要由α-螺旋组成,三维结构的基本特点是具有6个保守的α-螺旋,形成1个疏水性结合腔,这个结合腔即为CSPs的活性位点(徐浩智等,2015)。研究表明化学感受蛋白具有多种生理功能,包括:识别、结合并运输亲脂性的信息素分子(Pankiw et al., 2002),嗅觉功能(Monteforti et al., 2002),免疫功能(Sabatier et al., 2010),调节生理节律进而影响生长发育(Guo et al., 2011),信号转导(Calvello et al., 2005)。仁扇舟蛾(Clostera restitura)属于鳞翅目舟蛾科(Notodontidae)扇舟蛾属(Clostera),为杨树重要的食叶害虫。分布于我国的安徽、江苏、上海、浙江、江西、福建、湖南、广东、云南、海南、台湾和香港等地。国外分布于印度、越南、马来西亚以及印度尼西亚(武春生等,2003;刘群,2014a)。除了危害杨(Populus spp.),还会对柳(Salix spp.)及桦树(Betula spp.)造成危害。仁扇舟蛾1年发生6~7代(郑茂灿等,2006; 漆娟英等,2006;吴文杰等,2006),越冬卵翌年4月下旬开始孵化,初孵幼虫群集取食,3龄以后分散取食(刘群等,2014a)。仁扇舟蛾不仅世代多,并且极易暴发成灾。国内外关于仁扇舟蛾的研究着重于生物学特性及其与寄主植物相互关系(刘群,2014),但是关于仁扇舟蛾定位和寻找配偶及寄主植物的机制尚不清楚。本文通过研究仁扇舟蛾化学感受蛋白的cDNA的克隆、序列分析及组织表达特性,以期解析化学感受蛋白的功能,为阐明仁扇舟蛾在寻找配偶及寄主植物识别信息化学物质及其化学感受机制提供参考。
1 材料与方法 1.1 试验材料供试昆虫:仁扇舟蛾幼虫采自江苏省南京市浦口区乌江镇农田防护林的美洲黑杨(P.deltoides),带回实验室置于培养皿(直径150 mm,高20 mm)中,饲喂新鲜杨树叶子,在28 ℃、相对湿度为70%、光周期为12 h:12 h的条件下饲养。将1~5龄幼虫饥饿24 h后,置于RNA later中短时保存,通过离心法将RNAlater去除,转至液氮中速冻,置于-80 ℃保存备用。化蛹后,单头置于试管内,待成虫羽化后提供10%的蜂蜜水。成虫羽化后1~6天,选取大小相近,生理状态良好的雌雄虫各30头,分别解剖获取触角、头、足和翅,之后将组织样品同上述方法保存。
载体及菌种:大肠杆菌Trans1-T1hase Resistant Chemically Competent Cell、Peasy-T1载体购自北京全式金生物技术有限公司。
主要试剂和工具酶:总RNA提取试剂RNApure、UTaq DNA聚合酶购自北京庄盟国际生物基因科技有限公司;反转录试剂盒(PrimerScriptTMⅡ1st Strand Cdna Synthesis Kit)、RACE试剂盒(SMARTer® RACE 5′/3′ Kit User Manual)、荧光定量PCR试剂(SYBR® Premix Ex TaqTMⅡ)及程序均购自TaKaRa公司;DNA纯化回收试剂盒购自天根生化科技(北京)有限公司。
引物及测序:所用引物自行设计并由金斯瑞生物科技有限公司合成,质粒测序由上海杰李生物技术有限公司完成,转录组测序由北京华大生物科技有限公司测定。
1.2 仁扇舟蛾不同龄期幼虫、羽化后不同日龄及不同组织总RNA的提取及cDNA的合成分别选取仁扇舟蛾1~5龄幼虫虫体、羽化1~6日龄的雌、雄成虫触角,以及羽化3日龄成虫的头(不含触角)、触角、翅、足,液氮研磨后按照RNA提取试剂说明书提取总RNA,经电泳与分光度计检测后,根据TaKaRa公司反转录试剂盒说明书进行反转录获得cDNA。
1.3 仁扇舟蛾化学感受蛋白全长cDNA序列的克隆根据转录组数据,设计3′ RACE和5′ RACE引物(CSP-5′和CSP-3′)(表 1),以cDNA为模板,根据RACE试剂盒操作步骤进行3′和5′RACE的PCR扩增,将PCR产物经琼脂糖凝胶电泳后切胶纯化,克隆于Peasy-T1载体,导入大肠杆菌Trans1-T1之后在含有氨苄的并涂有IPTG及X-gal的LB琼脂板上37 ℃过夜培养,挑取阳性克隆于LB液体培养基中37 ℃、200 r·min-1孵育2 h,再进行菌液PCR鉴定。送样测序,将获得的测序片段进行拼接获得仁扇舟蛾CSP的全长cDNA序列。
|
|
qRT-PCR在Applied Biosystem 7500 System(USA)上进行。以小菜蛾ACTB(GeneBank:登录号AB282645)为内参基因,反应体系为20 μL:SYBR Premix Ex TaqⅡ10 μL,ROX Reference Dye 0.4 μL,上下游引物各0.8 μL(CSP-q-F和CSP-q-R,表 1),cDNA模板2 μL,dH2O 6 μL。反应条件95 ℃预变性30 s,40个循环:95 ℃ 5 s,60 ℃ 34 s,采用2-ΔΔCt方法计算Ct值,每个处理重复3次。
1.5 分析方法同源序列搜索使用在线工具(https://www.ncbi.nlm.nih.gov/),序列比对使用clustalx及DNAMAN软件,进化树构建使用MEGA6.0软件,信号肽预测利用在线网站(http://www.cbs.dtu.dk/services/SignalP/),等电点及分子质量预测利用在线网站(http://web.expasy.org/compute_pi/),蛋白三维结构的预测利用在线网站swiss model(http://www.swissmodel.expasy.org/)。荧光定量结果用2-Ct法计算相对表达量高低(Ct为循环阈值,表示每个反应中荧光信号到达设定阈值的循环数),ΔCt=目的基因平均Ct值-内参基因平均Ct值(Livak et al., 2001)。
2 结果与分析 2.1 仁扇舟蛾化学感受蛋白基因的全长克隆及序列分析通过RACE扩增并克隆获得仁扇舟蛾化学感受蛋白CresCSP3覆盖编码区的基因全长,序列分析表明CresCSP3覆盖编码区的cDNA全长为545 bp,包括61 bp的5′非翻译区,100 bp的3′非翻译区以及384 bp的开放阅读框,编码127个氨基酸,并且氨基酸序列中具有4个保守半胱氨酸位点,符合CX6CX17-19CX2C结构域,是典型的化学感受蛋白家族标志(图 1)。信号肽预测表明N末端含有18个氨基酸组成的信号肽,成熟蛋白预测分子质量14.34 kD,等电点5.75。

|
图 1 仁扇舟蛾化学感受蛋白基因CresCSP3的核苷酸序列及推导的氨基酸序列 Figure 1 The nucleotide and deduced amino acid sequences of CresCSP3 from the antennae of Clostera restitura 星号表示终止子, 方框所示4个保守的半胱氨酸, 下划线为预测的信号肽序列。The stop codon is indicated by an asterisk,conservative Cys sites are indicated by boxes and putative signal peptides are underlined. |
利用NCBI对化学感受蛋白进行检索,并列举CresCSP3与这12条CSP氨基酸序列的匹配度以及E值(表 2),CresCSP3与其他12种鳞翅目昆虫CSP序列同源性较低,与小菜蛾PxylCSP氨基酸序列相似度最高,为61%;与双委夜蛾、棉铃虫、亚洲玉米螟及甜菜夜蛾(Spodoptera exigua)的CSP氨基酸序列一致性分别为57%、57%、55%和55%,CresCSP3与茶尺蠖(Ectropis oblique hypulina)的EOblCSP2氨基酸序列一致性仅为27%。
|
|
应用Mega6.0软件Neighbor Joining(NJ)法对包括CresCSP3在内的13条CSP序列1 000次抽样分析来构建系统进化树(图 2),从系统发育树结果可以看出,CresCSP3并没有与其他种类聚为一支,说明CresCSP3与目前已有研究的其他鳞翅目昆虫CSP进化关系较远。

|
图 2 仁扇舟蛾化学感受蛋白CresCSP3与其他鳞翅目昆虫CSPs的系统发育树 Figure 2 The phylogenetic analysis of amino acid sequences of CresCSP3 in Clostera restitura and other lepidopteran species 图中的数值代表bootstrap为1 000次时的支持度值,标尺代表遗传距离。Bootstrap support values based on 1 000 replicates are indicated,and thescale bar indicates the phylogenetic distance. |
应用Swiss-model预测得到了CresCSP3编码蛋白三维结构,其三维结构与其他昆虫的CSP编码蛋白一样具有6个保守的α-螺旋,并形成1个疏水性的结合腔(图 3)。

|
图 3 预测的CresCSP3编码蛋白的三维结构 Figure 3 The predicted three-dimensional structure of CresCSP3 图中每种颜色代表一个α螺旋Each α-helix is indicated with a distinct color. |
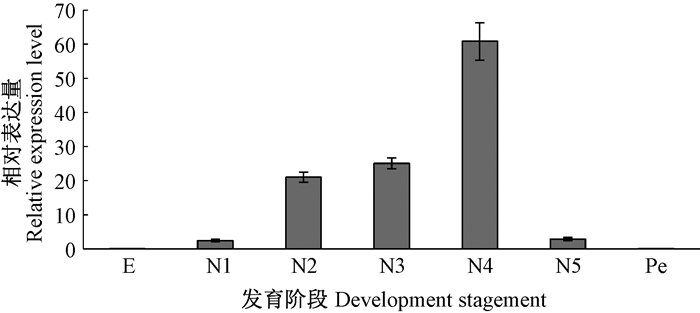
利用实时荧光定量PCR技术,分析CresCSP3在不同龄期幼虫、不同日龄成虫以及成虫不同组织间的表达差异,可知CresCSP3在不同龄期幼虫均有表达,在2~4龄期时表达量逐渐增加,并在4龄时表达量达到高峰(图 4)。羽化后雌、雄成虫1~6天均有表达,且都有1个表达高峰。雄虫在羽化后3、4天出现表达高峰,雌虫在羽化后4、5天出现表达高峰(图 5)。CresCSP3在4龄幼虫的相对表达量为62.28,而雄成虫4天时CresCSP3的相对表达量为132 475.86,雌成虫5天时相对表达量为93 798.02。在雌、雄成虫不同组织之间,CresCSP3在头部、触角、足及翅均有表达且在触角中的表达量最高。不同组织中CresCSP3相对表达量存在一定的性别差异,如在雄虫翅中的相对表达量明显高于雌虫(图 6)。
|
图 4 CresCSP3在仁扇舟蛾不同发育阶段的相对表达量 Figure 4 The relative expression level of CresCSP3 in C.restitura at different instars larvae E:卵Egg;N1:1龄幼虫1st instar larvae;N2:2龄幼虫2nd instar larvae;N3:3龄幼虫3rd instar larvae;N4:4龄幼虫4th instar larvae;N5:5龄幼虫5th instar larvae;Pe:蛹Pupae. |

|
图 5 CresCSP3在仁扇舟蛾成虫不同日龄的相对表达量 Figure 5 The relative expression level of CresCSP3 in adult C.restitura at different ages |

|
图 6 CresCSP3在羽化第3日成虫不同组织中的相对表达量 Figure 6 The relative expression level of CresCSP3 in adult C.restitura different tissues at the third day of eclosion |
仁扇舟蛾是杨树上的重要食叶害虫,深入了解其嗅觉感受机制,可以为开发新型引诱剂或趋避剂奠定基础。本研究采用RACE技术克隆获得仁扇舟蛾化学感受蛋白基因CresCSP3的cDNA全长,该cDNA编码127个氨基酸,有4个保守的半胱氨酸位点并且符合CX6CX17-19CX2C结构域,具有化学感受蛋白家族的典型特征。CresCSP3与其他鳞翅目昆虫CSP序列同源性较低,与小菜蛾PxylCSP氨基酸序列相似度最高,仅为61%,与双委夜蛾、棉铃虫、亚洲玉米螟及甜菜夜蛾的CSP氨基酸序列一致性分别为57%、57%、55%和55%,CresCSP3与茶尺蠖的EOblCSP2氨基酸序列一致性仅为27%。这些结果表明化学感受蛋白家族的多样性,不同物种的化学感受蛋白序列存在差异,可能影响蛋白的功能,进而出现功能行使上的差异,这与张国辉等(2012)对梨小食心虫化学感受蛋白的研究得出相似的结论。系统进化树分析表明,家蚕和野桑蚕、烟青虫和棉铃虫的CSP均以100%的置信度聚为同一分支,梨小食心虫和大螟CSP也以98%的置信度聚为一支,但CresCSP3没有与其他物种CSP聚为一支,说明CresCSP3基因与目前已有研究的其他鳞翅目昆虫CSP基因进化关系较远,可能与物种的分化时间有关,也可能由于相近物种CSP研究相对较少有关。
CresCSP3在卵期及蛹期表达量较低,幼虫1~5龄均有表达,2~4龄期随着取食量不断增加,表达量也逐渐增加,4龄时达到高峰,5龄时表达量略有降低,这与仁扇舟蛾在3龄取食量加大,末龄时取食量降低现象吻合(刘群等,2014),因此推断CresCSP3参与了仁扇舟蛾幼虫的取食过程。CresCSP3在不同日龄的雌、雄成虫触角上均有分布,但其表达情况有所不同。雄虫前4天CresCSP3的表达量高于雌虫,3~4天出现高峰,这可能是与雄虫感知性信息素寻找配偶的行为特性有关。雌雄成虫5天和4天时CresCSP3的相对表达量远高于4龄幼虫,表明仁扇舟蛾成虫需要表达大量的CresCSP3来进行交配和繁殖行为。这与对西花蓟马(Frankliniella occidentalis)中的化学感受蛋白的研究结果相似,其在成虫期的表达也高于幼虫,推测该基因很可能与其的性信息素的形成与释放、交尾等生殖系统的发育或行为有关(张治科等,2015)。另外,CresCSP3在雄虫翅中的表达量较高,交配时期,雌虫释放性信息素,雄虫会振翅寻找雌虫,推测其参与雄虫飞行过程中的气味识别与寻找配偶,进而证实CresCSP3参与雄虫交配行为和配偶定位。而雌虫羽化后4~5天为产卵高峰期,在其触角当中,4天之后CresCSP3的表达量的表达量升高,5天时达到高峰。雌虫交配之后需要寻找产卵位置,因此CresCSP3很可能参与雌虫产卵过程中的寄主定位。这与其他学者提出触角上的CSP具有寻找配偶及寄主定位的观点一致(Gu et al. 2012; Zhang et al. 2013)。
大多数气味结合蛋白于触角中特异性表达(Zhang et al., 2009),而化学感受蛋白不仅分布于昆虫触角(Liu et al., 2015),还分布于头、胸、腹、足、性腺等组织中(张国辉等,2012;郭兴国等,2016;Sun et al., 2016)。对家蚕的研究发现,化学感受蛋白存在于触角、喙、足、胸腔和腹部以及性腺中(Simona et al., 2006;Gong et al., 2007),本研究结果与其相似。刘金香等(2005)认为,昆虫化学感受蛋白广布于昆虫各组织,且蛋白质种类极丰富,这对于昆虫面对复杂的环境化学刺激信号而作出准确的行为反应具有重要意义。CresCSP3在雌、雄成虫的触角、头(不含触角)、足和翅中均有表达,在触角中表达量最高,因为昆虫主要通过触角感受外界挥发性气味分子,但对化学信号的识别除了触角外,头、足和翅等组织的化学感受器也有一定程度的参与。一般认为CSP的广泛分布与识别普通气味有关(谷少华等,2010),因此CresCSP3在仁扇舟蛾感知外界环境化学信号过程中具有重要作用。
4 结论本研究通过RACE技术获得仁扇舟蛾一个化学感受蛋白基因CresCSP3 cDNA序列,序列分析表明CSP家族的多样性,表明CSP在不同物种之间功能行使上的多样性;系统发育树分析说明CresCSP3基因与目前已有研究的其他鳞翅目昆虫CSP基因进化关系较远,可能与物种的分化时间有关,也可能由于相近物种CSP研究相对较少;成虫触角不同日龄的表达特性表明CresCSP3在仁扇舟蛾的寄主定位及配偶识别过程中具有重要作用,组织特异性表达特性也验证CresCSP3在仁扇舟蛾感知外界环境化学信号过程中扮演重要角色。本研究首次对仁扇舟蛾的化学感受蛋白CresCSP3进行相关研究,其功能尚需进一步验证,仁扇舟蛾其他化学感受蛋白基因有待挖掘和深入研究。
邓培渊, 袁伟, 李玉华, 等. 2015. 甜菜夜蛾化学感受蛋白SexiCSP3结合特性分析[J]. 环境昆虫学报, 37(5): 979-986. (Dng P Y, Yuan W, Li Y H, et al. 2015. Binding specificity of the chemosensory protein SexiCSP3 in the Beet Armyworm, Spodoptera exigua (Hübner)[J]. Journal of Environmental Entomology, 37(5): 979-986. [in Chinese]) |
谷少华, 张雪莹, 张永军, 等. 2010. 苜蓿盲蝽气味结合蛋白基因Alin-OBP1的克隆及表达谱分析[J]. 昆虫学报, 53(5): 487-496. (Gu S H, Zhang X Y, Zhang Y J, et al. 2010. Cloning and expression pattern analysis of an odorant binding protein gene Alin-OBP1 in the lucerne plant bug, Adelphocoris lineolatus(Goeze)(Hemiptera:Miridae)[J]. Acta Entomologica Sinica, 53(5): 487-496. [in Chinese]) |
郭兴国, 陈莹, 邢秋婷, 等. 2016. 家蚕化学感受蛋白CSP16的表达及结合特性分析[J]. 昆虫学报, 59(6): 613-621. (Guo X G, Chen Y, Xing Q T, et al. 2016. Expression and binding characterization of chemosensory protein CSP16 in the silkworm, Bombyx mori[J]. Acta Entomologica Sinica, 59(6): 613-621. [in Chinese]) |
李红亮, 倪翠侠, 姚瑞, 等. 2010. 中华蜜蜂化学感受蛋白基因Acer-CSP1克隆与表达特征分析[J]. 昆虫学报, 53(9): 962-968. (Li H L, Ni C X, Yao R, et al. 2010. Molecular cloning, characterization, and expression pattern of chemosensory protein 1 gene (Acer-CSP1) in the Chinese honeybee, Apisceranacerana (Hymenoptera:Apidae)[J]. Acta Entomologica Sinica, 53(9): 962-968. [in Chinese]) |
刘群, 常虹, 陈娟, 等. 2014. 分月扇舟蛾与仁扇舟蛾的形态学和生物学区别及其进化关系[J]. 林业科学, 50(1): 97-102. (Liu Q, Chang H, Chen J, et al. 2014. Identification of Clostera anastomosis and C.restitura (Lepidoptera:Notodontidae) by morphological and biological characters and their evolutionary[J]. Scientia Silvae Sinicae, 50(1): 97-102. [in Chinese]) |
刘群. 2014. 仁扇舟蛾的形态学及其诱导杨树抗性文库的构建. 南京: 南京林业大学硕士学位论文. (Liu Q. 2014. Morphological characteristics of Clostera restitura and the construction of the induced full-length cDNA library of Populus deltoids by C. restitura feeding. Nanjing: MS thesis of Nanjing Forestry University. [in Chinese]) |
刘金香, 钟国华, 谢建军, 等. 2005. 昆虫化学感受蛋白研究进展[J]. 昆虫学报, 48(3): 418-426. (Liu J X, Zhong G H, Xie J J, et al. 2005. Recent advances in chemosensory proteins of insects[J]. Acta Entomologica Sinica, 48(3): 418-426. [in Chinese]) |
马双, 王松莹, 谷少华, 等. 2011. 苜蓿盲蝽化学感受蛋白Alin-CSP6的气味结合特征[J]. 中国农业科学, 44(14): 2926-2934. (Ma S, Wang S Y, Gu S H, et al. 2011. Odorant binding characteristics of chemosensory protein Alin-CSP6 in Lucerne Plant Bug, Adelphocorislineolatus (Goeze)[J]. Scientia Agricultura Sinica, 44(14): 2926-2934. DOI:10.3864/j.issn.0578-1752.2011.14.009 [in Chinese]) |
王桂荣, 郭予元, 吴孔明. 2002. 棉铃虫触角感器的超微结构观察[J]. 中国农业科学, 35(12): 1479-1482. (Wang G R, Guo Y Y, Wu K M. 2002. Observation on the ultrastructures of antennal sensilla in Helicoverpa armigera[J]. Scientia Agricultura Sinica, 35(12): 1479-1482. DOI:10.3321/j.issn:0578-1752.2002.12.008 [in Chinese]) |
王思豹, 张帅, 雒珺瑜, 等. 2015. 大草蛉化学感受蛋白基因CpalCSP3组织表达谱及气味结合特性分析[J]. 棉花学报, 27(3): 260-267. (Wang S B, Zhang S, Luo J Y, et al. 2015. Tissue expression profile and ligand binding affinity of the chemosensory protein gene CpalCSP3 in Chrysopapallens(Rambur)[J]. Cotton Science, 27(3): 260-267. DOI:10.11963/issn.1002-7807.201503010 [in Chinese]) |
武春生, 方承莱. 2003. 中国动物志. 昆虫纲. 第三十一卷. 鳞翅目: 舟蛾科. 北京: 科学出版社. (Wu C S, Fang C L. 2003. Fauna sinica. Isecta. Volume 31. Lepidoptera: Notodontidae. Beijing: Science Press. [in Chinese]) |
徐浩智, 游银伟, 张龙. 2015. 昆虫化学感受蛋白及其功能研究进展[J]. 农业生物技术学报, 23(01): 118-125. (Xu H Z, You Y W, Zhang L. 2015. A review on chemosensory protein and its functions in insects[J]. Journal of Agricultural Biotechnology, 23(1): 118-125. [in Chinese]) |
张国辉, 刘彦飞, 仵均祥. 2012. 梨小食心虫化学感受蛋白cDNA的克隆、序列分析及原核表达[J]. 昆虫学报, 55(6): 668-675. (Zhang G H, Liu Y F, Wu J X. 2012. cDNA cloning, sequence analysis and prokaryotic expression of a chemosensory protein from the oriental fruit moth, Grapholitamolesta(Lepidoptera:Tortricidae)[J]. Acta EntomologicaSinica, 55(6): 668-675. [in Chinese]) |
张天涛, 邹朗云, 李科明, 等. 2011. 棉铃虫化学感受蛋白Harm CSP6二聚体的组织表达分析及气味结合特征[J]. 昆虫学报, 54(6): 615-622. (Zhang T T, Zou L Y, Li K M, et al. 2011. Expression profiling and binding characterization of dimeric chemosensory protein 6(Harm CSP6) in Helicoverpaarmigera (Hübner)(Lepidoptera:Noctuidae)[J]. Acta Entomologica Sinica, 54(6): 615-622. [in Chinese]) |
张志春, 王满囷, 张国安. 2009. 小菜蛾化学感受蛋白基因PxylCSP1的克隆和表达[J]. 昆虫学报, 52(2): 140-146. (Zhang Z C, Wang M Q, Zhang G A. 2009. Molecular cloning and expression characterization of a chemosensory protein gene PxylCSP1 from the diamondback moth, Plutellaxylostella(Lepidoptera:Plutelidae)[J]. Acta Entomologica Sinica, 52(2): 140-146. [in Chinese]) |
张治科, 吴圣勇, 雷仲仁. 2015. 西花蓟马化学感受蛋白的cDNA克隆、时空表达分析及组织定位[J]. 昆虫学报, 58(1): 1-14. (Zhang Z K, Wu S Y, Lei Z R. 2015. cDNA cloning, expression profiling and immunolocalization of a chemosensory protein in the western flower thrips, Frankliniella occidentalis(Thysanoptera:Thtipidae)[J]. Acta Entomologica Sinica, 58(1): 1-14. DOI:10.7679/j.issn.2095-1353.2015.001 [in Chinese]) |
赵洁, 陆永跃. 2015. 棉花粉蚧化学感受蛋白基因的鉴定与进化分析[J]. 广东农业科学, 12(13): 121-127. (Zhao J, Lu Y Y. 2015. Identification and evolution analysis on chemosensory protein genes in Phenacoccus solenopsis Tinsley (Hemiptera:Pseudococcidae)[J]. Guangdong Agricultural Sciences, 12(13): 121-127. [in Chinese]) |
Brito N F, Moreira M F, Melo A C. 2016. A look inside odorant-binding proteins in insect chemoreception[J]. Journal of Insect Physiology, 95(12): 51-65. |
Calvello M, Brandazzaa A, Navarrinia A, et al. 2005. Expression of odorant-binding proteins and chemosensory proteins in some Hymenoptera[J]. Insect Biochemistry & Molecular Biology, 35(4): 297-307. |
Gong D P, Zhang H J, Zhao P, et al. 2007. Identification and expression pattern of the chemosensory protein gene family in the silkworm, Bombyx mori[J]. Insect Biochemistry & Molecular Biology, 37(3): 266-277. |
Gu S H, Wang S Y, Zhang X Y, et al. 2012. Functional characterizations of chemosensory proteins of the alfalfa plant bug Adelphocoris lineolatus indicate their involvement in host recognition[J]. Plos One, 7(8): e42871. |
Guo W, Wang X, Ma Z, et al. 2011. CSP and takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust[J]. PLoS Genet, 7(2): e1001291. |
Liu Q, Chen J, Hao D J. 2013. Scanning electron microscopy studies of antennal sensilla in Clostera anastomosis Linnaeus (Lepidoptera:Notodontidae)[J]. Entomological News, 123(2): 110-130. |
Liu S, Shi X X, Zhu Q Z, et al. 2015. Identification and expression profiles of putative chemosensory protein genes in Cnaphalocrocis medinalis (Lepidoptera:Pyralidae)[J]. Journal of Asia-Pacific Entomology, 18(1): 99-105. |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) Method[J]. Methods, 25(4): 402-408. |
Marchese S, Angeli S, Andolfo A, et al. 2000. Soluble proteins from chemosensory organs of Eurycantha calcarata(Insect, Phasmatodea)[J]. Insect Biochem Molec Biol, 30(11): 1091-1098. |
Mckenna M P, Hekmatscafe D S, Gaines P, et al. 1994. Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system[J]. Journal of Biological Chemistry, 269(23): 16340-16347. |
Monteforti G, Angeli S, Petacchi R, et al. 2002. Ultrastructural characterization of antennal sensilla and immunocytochemical localization of a chemosensory protein in Carausius morosus Brunner (Phasmida:Phasmatidae)[J]. Arthropod Structure & Development, 30(3): 195-205. |
Pankiw T, Rubink W. 2002. Pollen foraging response to brood pheromone by Africanized and European honey bees (Apis mellifera L.)[J]. Annals of the Entomological Society of America, 95(6): 761-767. |
Pelletier J, Leal W S. 2011. Characterization of olfactory genes in the antennae of the Southern house mosquito, Culexquinque fasciatus[J]. Inect Physiology, 57(7): 915-929. |
Pelosi P, Iovinella I, Felicioli A, et al. 2014. Soluble proteins of chemical communication:an overview across arthropods[J]. Frontiers in Physiology, 5(8): 320. |
Pelosi P, Zhou J J, Ban L P, et al. 2006. Soluble proteins in insect chemical communication[J]. Cellular & Molecular Life Sciences Cmls, 63(14): 1658-1676. |
Picimbon J F, Dietrich K, Breer H, et al. 2000. Chemosensory proteins of Locusta migratoria (Orthoptera:Acrididae)[J]. Insect Biochemistry & Molecular Biology, 30(3): 233-241. |
Picimbon J F, Leal W S. 1999. Olfactory soluble proteins of cockroaches[J]. Insect Biochemistry & Molecular Biology, 29(11): 973-978. |
Sabatier L, Jouanguy E, Dostert C, et al. 2010. Two drosophila molecules related to pheromone/odor-binding proteins induced by viral and bacterial infections[J]. European Journal of Biochemistry, 270(16): 3398-3407. |
Tomaselli S, Crescenzi O, Sanfelice D, et al. 2006. Solution structure of a chemosensory protein from the desert locust Schistocerca gregaria[J]. Biochemistry, 45(35): 10606-10613. |
Steinbrecht R A, Laue M, Ziegelberger G. 1995. Immunolocalization of pheromone-binding protein and general odorant-binding protein in olfactory sensilla of the silk moths Antheraea and Bombyx[J]. Cell and Tissue Research, 282(2): 203-217. DOI:10.1007/BF00319112 |
Sun H, Song Y, Du J, et al. 2016. Identification and tissue distribution of chemosensory protein and odorant binding protein genes in Athetisdis similis (Lepidoptera:Noctuidae)[J]. Applied Entomology and Zoology, 51(3): 409-420. DOI:10.1007/s13355-016-0413-8 |
Vogt R G, Miller N E, Litvack R, et al. 2009. The insect SNMP gene family[J]. Insect Biochemistry & Molecular Biology, 39(7): 448-456. |
Vogt R G, Prestwich G D, Lerner M R. 1991. Odorant-binding-protein subfamilies associate with distinct classes of olfactory receptor neurons in insects[J]. Journal of Neurobiology, 22(1): 74-84. DOI:10.1002/(ISSN)1097-4695 |
Vogt R G, Riddiford L M, Pestwich G D. 1985. Kinetic properties of a sex pheromone-degrading enzyme:the sensillar esterase of Antheraea Polyphemus[J]. Proceedings of the National Academy of Sciences, 82(24): 8827-8831. DOI:10.1073/pnas.82.24.8827 |
Xu Y L, He P, Zhang L, et al. 2009. Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects[J]. BMC Genomics, 10(1): 632. DOI:10.1186/1471-2164-10-632 |
Yang B, Ozaki K, Ishikawa Y, et al. 2016. Sexually biased expression of odorant-binding proteins and chemosensory proteins in Asian corn borer Ostrinia furnacalis (Lepidoptera:Crambidae)[J]. Applied Entomology and Zoology, 51(3): 373-383. DOI:10.1007/s13355-016-0409-4 |
Zhang J, Wang B, Dong S, et al. 2015. Antennal transcriptome analysis and comparison of chemosensory gene families in two closely related noctuidae moths, Helicoverpa armigera and H. assulta[J]. Plos One, 10(2). |
Zhang Y N, Ye Z F, Yang K, et al. 2013. Antenna-predominant and male-biased CSP19 of Sesamia inferens, is able to bind the female sex pheromones and host plant volatiles[J]. Gene, 536(2): 279-286. |
Zhang Z C, Wang M Q, Lu Y B, et al. 2009. Molecular characterization and expression pattern of two general odorant binding proteins from the diamondback moth, Plutella xylostella[J]. Journal of Chemical Ecology, 35(10): 1188. DOI:10.1007/s10886-009-9697-2 |
 2018, Vol. 54
2018, Vol. 54

