文章信息
- 李雯雯, 赵文霞, 林若竹, 姚艳霞, 李娟, 淮稳霞
- Li Wenwen, Zhao Wenxia, Lin Ruozhu, Yao Yanxia, Li Juan, Huai Wenxia
- 基于ITS和β-tubulin基因分析的居间疫霉菌系统发育
- Phylogenetic Relationships of Phytophthora intercalaris Based on ITS and β-tubulin Gene Sequence
- 林业科学, 2018, 54(3): 73-82.
- Scientia Silvae Sinicae, 2018, 54(3): 73-82.
- DOI: 10.11707/j.1001-7488.20180308
-
文章历史
- 收稿日期:2017-08-17
- 修回日期:2017-10-23
-
作者相关文章
2. 国家林业局森林病虫害防治总站 沈阳 110034
2. Forest Pest Control Station of State Forestry Administration Shenyang 110034
疫霉菌(Phytophthora)属于卵菌门(Oomycota)卵菌纲(Oomycetes)腐霉目(Pythiales)腐霉科(Pythiaceae)(Cavalier-Smith, 2004),是一类重要的植物病原菌,寄主范围较广,可侵染林木、果树、农作物和园林花卉等多种植物,引起枝枯、腐烂、溃疡、猝倒等症状,严重时导致全株死亡,可给森林生态系统和农业生产带来巨大的危害和损失(郑小波,1997)。有研究表明,疫霉菌属的有些种能够严重危害森林生态系统,例如栎树猝死病菌(Phytophthora ramorum)在欧美多个国家引起毁灭性的森林病害(Werres et al., 2001; Rizzo et al., 2005; Brasier et al., 2012),对林产品贸易造成巨大的损失,被我国等多个国家列入进境植物检疫名单。2007年在美国俄勒冈州森林中首次发现的锡斯基尤疫霉菌(P. siskiyouensis)可以造成石栎(Lithocarpus densiflorus)、加州月桂(Umbellularia californica)和桤木(Alnus spp.)等多种树木枝干产生溃疡,严重时可致死(Reeser et al., 2007)。
疫霉菌危害的严重性已引起国内外众多专家学者的重视,随着分子生物学技术的迅速发展以及调查范围的扩大,该属内越来越多的种被发现,从20世纪90年代的58个种到目前的增加至超过130个种(Kroon et al., 2012; Martin et al., 2012, 2014; Roy et al., 2012; Yang et al., 2016; Brazee et al., 2016)。Cooke等(2000)根据ITS序列分析,将疫霉菌属划分为8个进化枝,之后Blair等(2008)通过多基因系统发育分析又增加了2个进化枝,将疫霉菌属划分为10个进化枝。中国目前报道疫霉属有57个种,其中包括7个正式描述的新种和8个未正式描述的种(郑小波,1997;余永年,1998;Zeng et al., 2009; Huai et al., 2013; Jung et al., 2017a;2017b; Li et al., 2017)。我国对于疫霉菌的研究多集中在农作上,如大豆疫霉菌(P. sojea)、辣椒疫霉菌(P. capsici)等(孙文秀,2005; 张正光等,2003; 朱振东等,2006)的遗传和重要功能基因的研究以及防治等方面,而对于森林中的疫霉菌以及疫霉病害研究相对比较薄弱。我国地域辽阔,气候类型复杂,森林类型多样,疫霉菌种类以及遗传多样性可能更丰富。笔者自2005年起,在全国范围内展开关于森林疫霉菌的调查,并在我国西南地区杜鹃(Rhododendron)-栎(Quercus)树林中的溪流和土壤中发现了8个新记录种(Huai et al., 2013),这不仅扩大了疫霉菌种群的分布范围,也增加了我国疫霉菌的种类。笔者在2013年对四川省西昌市的泸山森林公园进行调查时,发现了疫霉菌新记录种居间疫霉菌(Phytophthora intercalaris),并对其形态特征和分子系统发育做了初步研究,以期为研究疫霉菌相关的林木衰退病的发病规律打下基础,为制定林木疫病的防治策略提供理论支持。
1 材料与方法 1.1 菌株的采集与分离在四川省西昌市泸山的溪流中利用大叶榕(Ficus altissima)、小叶榕(F. concinna)和青冈(Cyclobalanopsis glauca)的健康叶片诱捕疫霉菌,然后进行疫霉菌株分离。取3~5片健康叶片放入网袋内,然后将其浸泡在监测点的溪流里,用绳子将网袋一端固定。7天之后取出诱捕叶片,重新放入健康叶片。取出的叶片先用清水洗干净晾干,然后用75%的酒精擦拭表面,在病健交界处切取植物组织(约4 mm×2 mm),置于选择性培养基CARP+(17 g玉米粉培养基,1 L蒸馏水,20 mg匹马霉素,200 mg氨苄青霉素,10 mg利福霉素,30 mg苯菌灵,50 mg恶霉灵)上,室温下培养3~5天。待组织边缘长出菌丝后,置于显微镜下观察,挑取疫霉菌丝转入选择性培养基CARP(与选择性培养基CARP+相比无苯菌灵和恶霉灵2种抗生素),进一步纯化,待菌落长好后转入玉米粉培养基(CMA)保藏。将纯菌株接种到V8固体培养基上培养4~5天,用直径6 mm的打孔器沿菌落边缘打孔,将菌丝块转入空培养皿中,加入无菌水至没过菌丝块,诱导其产孢。室温日光下培养1天,置于显微镜(Zeiss Primo Star, Carl Zeiss)下观察并用CCD相机(TCC-5.0,图森图像技术有限公司)拍照。
1.2 DNA提取及PCR扩增在装有菌丝块的2 mL离心管中,加入2颗玻璃珠及少量石英砂,在震荡仪上震荡研磨10 min。再用改良的CTAB法(Huai et al., 2003)提取DNA。ITS序列扩增正向引物为ITS6(5′-GAAGGTGA AGTCGTAACAAGG-3′) (Cooke et al., 2000),反向引物为ITS4(5′-TCCTCCGCTTATTGATATGC-3′)(White et al., 1990)。β-tubulin序列扩增引物为TUBUF2(5′-CGGTAACAACTGGGCCAAGG-3′)(Kroon et al., 2004)和TUBUR1(5′-CCTGGTACTGCTGGTACTCAG-3′)(Kroon et al., 2004)。25 μL的PCR反应体系包括:DNA模板1.5 μL,10 μmol ·L-1 ITS引物各0.75 μL (β-tubulin引物各0.25 μL),2×Taq PCR MasterMix 12.5 μL,ddH2O 9.5 μL(β-tubulin体系中ddH2O 10.5 μL)。ITS序列扩增程序为:94 ℃ 5 min;94 ℃ 1 min,58 ℃ 30 s, 72 ℃ 1 min, 35个循环;72 ℃ 10 min,24 ℃ 1 min。β-tubulin序列扩增程序为:94 ℃ 2 min;94 ℃ 30 s,58 ℃ 30 s,72 ℃ 1 min,35个循环;72 ℃ 10 min。PCR产物由美吉生物科技有限公司进行双向测序。
1.3 序列分析及系统发育测序结果用Staden Package 1.6.0进行拼接,然后将其与GenBank中已有序列进行比对(https://blast.ncbi.nlm.nih.gov/Blast.cgi),选择下载相似度高且已发表的序列进一步分析(表 1)。用MAFFT version 7(Katoh et al., 2013)进行在线序列比对, 比对后的序列用贝叶斯方法(Bayesian)、最大简约法(MP)和最大似然法(ML)进行系统发育分析,构建系统发育树。利用贝叶斯法建树时,先用Modeltest 2.3软件选择最佳模型,然后在MrBayes 3.2.6中采用Markov Chain Monte Carlo(MCMC)算法,运行1 000 000代。利用PAUP version 4.0 beta 10(Swofford, 2002)构建MP树时,采用启发式搜索(heuristic search)和树二重组(TBR)交换算法,序列排列导致的空位(gap)被视作碱基缺失。所有碱基权重相等,计算树长(tree length, TL)、一致性指数(consistency index,CI)、相似性指数(homoplasy index, HI)、保留指数(retention index, RI)和校正一致性指数(rescaled consistency index, RC)。利用PhyML 3.0在线构建ML树,选择GTR模型。另外,在构建ITS和β-tubulin系统发育树时,选择腐霉属的Pythium aphanidermatum作为外群。建好的Bayesian树和ML树用FigTree version 1.4.0软件查看。
|
|
试验分离得到了46个菌株, 显微镜下初步观察,形态特征与居间疫霉菌相似(Yang et al., 2016)。居间疫霉菌在无菌水中诱导20 h左右产生大量的孢子囊,多卵形,偶有倒梨形,端生,无乳突,不脱落,内层出。在固体V8培养基上几乎不产生菌丝膨大体。在无菌水中产生大量薄壁的厚垣孢子,多间生,也有侧生。异宗配合,单个菌株培养不产生藏卵器和雄器,如图 1所示。

|
图 1 居间疫霉菌的形态结构特征 Figure 1 Morphological characteristics of P. intercalaris a.卵圆形孢子囊Ovoid sporangia; b.近球形孢子囊Nearly spherical sporangia; c.孢子囊内层出Extended proliferation; d.菌丝膨大体Hyphal swelling; e, f.侧生厚垣孢子Lateral chlamydospore; g.夹生厚垣孢子Intercalary chlamydospore; h.着生于小枝的厚垣孢子Chlamydospore on short branch. |
经过拼接之后,所获得的46株疫霉菌的完整ITS序列为847~849 bp,且彼此间仅有1~3个碱基的差异(表 2),序列一致性为99.65% ~99.88%,与参考居间疫霉菌菌株(KT163268)的序列一致性为99.29% ~99.53%。将7个代表菌株的序列与38条参比序列用MAFFT比对之后,构建最大简约树(MP),分析结果表明,969个序列特征无序且权重相等,其中衡量特征431个,简约无信息的变量特征135个,简约信息特征403个。MP树的树长为1 931,一致性指数为0.481 1,相似性指数为0.518 9,保留指数为0.746 7,校正一致性指数(RC)为0.359 2。最大简约法、最大似然法和贝叶斯法构建的系统发育树虽然在末端分支模式和分支长度上有细微差异,但3种树的拓扑结构基本一致。如图 2所示,7个代表菌株均与居间疫霉菌聚为一枝,支持率为100%。MP树和ML树的节点支持率以及贝叶斯树的后验概率如图 2所示。
|
|

|
图 2 基于ITS序列构建的疫霉菌系统发育树 Figure 2 Phylogenetic tree of Phytophthora based on ITS sequence 图中数字分别依次表示贝叶斯后验概率、MP树及ML树的节点支持率,低于60%的未显示。 Nodes of support values of Bayesian tree, MP tree and ML tree were shown on the phylogenetic tree, and the values less than 60% were not shown. |
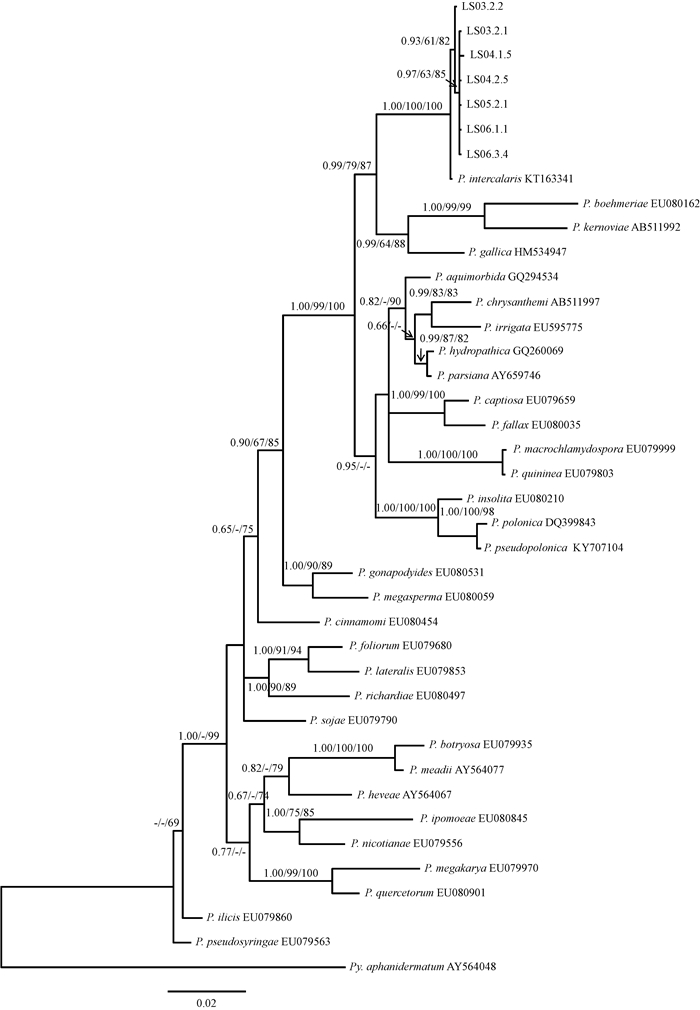
拼接之后所获得的40株疫霉菌的完整β-tubulin序列均为882 bp,且彼此间仅有1~2个碱基的差异(表 3),序列一致性为99.77% ~99.89%,与参考居间疫霉菌菌株(KT163336)的序列一致性为99.43% ~99.66%。将7个代表菌株的序列与33条参比序列用MAFFT比对之后,构建最大简约树(MP),分析结果表明,882个序列特征无序且权重相等,其中衡量特征640个,简约无信息的变量特征63个,简约信息特征179个。MP树的树长为866,一致性指数为0.401 8,相似性指数为0.598 2,保留指数为0.613 7,校正一致性指数为0.246 6。由图 3可知,供试菌株以100%的支持率全部与居间疫霉菌聚为一支,但菌株序列之间存在碱基差异, 菌株LS03.2.2的序列与其他序列差异相对较大,所以在居间疫霉菌这一大的分支下又单独形成一个小支,且支持率也比较高。MP树和ML树的节点支持率以及贝叶斯树的后验概率如图 3所示。
|
|
|
图 3 基于β-tubulin序列构建的疫霉菌系统发育树 Figure 3 Phylogenetic tree of Phytophthora based on β-tubulin sequence 图中数字分别依次表示贝叶斯后验概率、MP树及ML树的节点支持率,低于60%的未显示。 Nodes of support values of Bayesian tree, MP tree and ML tree were shown on the phylogenetic tree, and the values less than 60% were not shown. |
本研究通过亚热带部分林间溪流里的疫霉菌的分离、观察和基于ITS和β-tubulin基因位点的系统发育分析证实:试验分离到的46株疫霉菌均为居间疫霉菌,是我国的新记录种。
居间疫霉菌最早发现于美国东部地区,是在林间溪流里诱捕到的(Yang et al., 2016)。基于ITS和COXI基因的系统发育结果显示其属于clade10,与瘿疫霉菌(P. gallica)亲缘关系很近(Yang et al., 2016)。虽然二者在形态上都产生无乳突、不脱落的孢子囊,但是在有性生殖方面差异较大,前者是异宗配合,A1交配型;后者是自身不育,单独培养或者与A1型和A2型分别配对均不产生配子囊(Yang et al., 2016;Jung et al., 2008)。另外,前者是在溪流中分离到的,后者是在衰弱木的根际土壤中发现的(Yang et al., 2016;Jung et al., 2008)。在系统发育上同属clade10的苎麻疫霉菌、康沃尔疫霉菌和番薯疫霉菌,与居间疫霉菌在形态上存在较大差异,它们在Waterhouse的形态学分组上属于Group Ⅱ,同宗配合,产生的孢子囊有乳突、易脱落(Brasier et al., 2005; Erwin et al., 1996; Nelson et al., 2010)。Jung等(2011)认为卵孢子在自然选择对疫霉菌有性生殖的影响方面起着至关重要的作用,异宗配合形成的卵孢子不仅能够通过重组形成新的基因型,还可以作为休眠结构度过干旱、高温、低温等不良环境;同宗配合产生的卵孢子可以提供休眠结构,但是重组和对环境改变的适应能力受到限制。据此笔者推测,居间疫霉菌遵循此进化途径,其在所处的生态位中是优势种群。目前,居间疫霉菌仅在美国发现,其他国家和地区并未见报道,本研究在四川省西昌市分离到该菌,扩大了其分布范围。
关于疫霉菌鉴定和分类,专家学者已经做了大量的研究(Cooke et al., 1997; Forster et al., 2000; Martin et al., 2003; Kroon et al., 2004; Uddin et al., 2007),结果表明疫霉菌不同种的ITS序列具有较大的多态性。Xu等(2007)的研究表明不同地理来源的大豆疫霉菌菌株的ITS序列差异较大,而柑橘褐腐疫霉菌(P.citrophthora)、P. quercina和P. megasperma的系统发育分析则表明同一种内来自不同地域和寄主的菌株差异并不大(Uddin et al., 2007; Balci et al.; 2003;Forster et al., 1993)。Martin等(2004)的研究也表明疫霉菌的很多种都极少存在种内多态性。本研究首次将不同地理来源的居间疫霉菌做了基于rDNA-ITS与β-tubulin序列的系统发育分析,结果显示中国和美国的菌株以较高的支持率聚为一支,不同菌株间序列一致性很高,但也存在一些碱基位点差异,造成这种差异的原因:一是存在种间杂交,居间疫霉菌是异宗配合,杂交可能会导致种内或者个体内的基因位点的变异,通过重组形成新的基因型(Jung et al., 2011);二是单个核苷酸位点上存在变异,DNA在复制的时候可能会发生碱基对的替换、增添和缺失,这就导致了序列位点的差异,然而由于简并碱基的存在,使变异有时候并不会引起性状的改变。所以,导致居间疫霉菌菌株间序列差异的具体原因还有待进一步的研究。
疫霉菌是一种重要的植物病原菌,不同种类的疫霉菌在致病性、寄主等方面存在很大差异,在生态系统中的作用也值得深入研究。樟疫霉菌分布广泛,全世界70多个国家和地区都有报道,能够侵染273属900多种寄主植物(余永年,1998),在森林生态系统中造成易感植物种群丰富度下降,同时也使得植物群落结构和野生抗性物种生物量发生改变(Wills, 1992)。节水霉状疫霉菌不仅是很多植物的病原菌,可能还在生态系统中扮演着重要角色,分解绿色植物的凋落物和碎屑,这使得节水霉状疫霉菌在自然生态系统中广泛存在(Brasier et al., 2003)。很多土壤或者水域环境中的疫霉菌往往不会直接造成严重的病害,必须是在环境条件高度适合的情况下才可能发生(Brasier et al., 2003)。居间疫霉菌几乎所有的菌株都是在溪流里分离得到的,仅有1株是在农田灌溉水中发现(Yang et al., 2016),但这还不足以说明居间疫霉菌能够适应农田生态系统。另外,植物组织和根际土壤中是否存在尚不可知,目前也并没有发现与之直接相关的林木病害,居间疫霉菌是否在森林衰退病中起到一定作用还有待研究,致病性还有待明确。
4 结论对中国新记录种居间疫霉菌种群的系统发育研究表明,其ITS序列为847~849 bp,菌株间仅有1~3个碱基差异,与参考菌株(KT163268)序列一致性为99.29% ~99.53%;β-tubulin序列均为882 bp,菌株间仅有1~2个碱基差异,与参考菌株(KT163336)序列一致性为99.43% ~99.66%。系统发育分析结果显示,不同地理来源的居间疫霉菌种群序列一致性很高,存在单个碱基位点的变异,形成多个不同的基因型。本研究不仅增加了居间疫霉菌的菌株数量,也扩大了该菌的分布范围,同时也丰富了中国疫霉菌的种类。
孙文秀, 张修国, 贾永健, 等. 2005. 不同地区辣椒疫霉菌遗传多样性的RAPD分析[J]. 植物病理学报, 35(4): 340-344. (Sun W X, Zhang X G, Jia Y J, et al. 2005. Phylogenetic analysis among isolates of Phytophthora capsici from different areas by RAPD[J]. Acta Phytopathologica Sinica, 35(4): 340-344. [in Chinese]) |
余永年. 1998. 中国真菌志.第6卷.霜霉目[M]. 北京: 科学出版社. (Yu Y N. 1998. Chinese Fungi Record.Vol.6. Peronosporales[M]. Beijing: Science Press. [in Chinese]) |
张正光, 王源超, 郑小波. 2003. 大豆疫霉和苜蓿疫霉rDNA ITS序列分析[J]. 菌物学报, 22(4): 542-548. (Zhang Z G, Wang Y C, Zheng X B. 2003. Analysis of rDNA ITS sequence of Phytophthora sojae and P. medicaginis[J]. Mycosystema, 22(4): 542-548. [in Chinese]) |
郑小波. 1997. 疫霉菌及其研究技术[M]. 北京: 中国农业出版社. (Zheng X B. 1997. Phytophthora and its research technique[M]. Beijing: China Agriculture Press. [in Chinese]) |
朱振东, 霍云龙, 王晓鸣, 等. 2006. 大豆疫霉根腐病抗源筛选[J]. 植物遗传资源学报, 7(1): 24-30. (Zhu Z D, Huo Y L, Wang X M, et al. 2006. Screening for resistance sources to Phytophthora root rot of soybean[J]. Journal of Plant Genetic Resources, 7(1): 24-30. [in Chinese]) |
Balci Y, Halmschlager E. 2003. First report of Phytophthora quercina from oak forests in Austria[J]. Plant Pathology, 52(3): 403-403. DOI:10.1046/j.1365-3059.2003.00825.x |
Blair J E, Coffey M D, Park S Y, et al. 2008. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences[J]. Fungal Genetics and Biology, 45(3): 266-277. DOI:10.1016/j.fgb.2007.10.010 |
Brasier C M, Beales P A, Kirk S A, et al. 2005. Phytophthora kernoviae sp.nov. an invasive pathogen causing bleeding stem lesions on forest trees and foliar necrosis of ornamentals in the UK[J]. Mycological Research, 109(8): 853-859. DOI:10.1017/S0953756205003357 |
Brasier C M, Cooke D E, Duncan J M, et al. 2003. Multiple new phenotypic taxa from trees and riparian ecosystems in Phytophthora gonapodyides-P. megasperma ITS clade 6, which tend to be high-temperature tolerant and either inbreeding or sterile[J]. Mycological Research, 107(3): 277-299. DOI:10.1017/S095375620300738X |
Brasier C M, Webber J F. 2012. Natural stem infection of Lawson cypress (Chamaecyparis lawsoniana) caused by Phytophthora ramorum[J]. New Disease Report, 25: 26. DOI:10.5197/j.2044-0588.2012.025 |
Brazee N J, Wick R L, Hulvey J P. 2016. Phytophthora species recovered from the Connecticut river valley in Massachusetts, USA[J]. Mycologia, 108(1): 6-19. DOI:10.3852/15-038 |
Cavalier-Smith T. 2004. Only six kingdoms of life[J]. Proceedings of the Royal Society B, 271: 1251-1262. DOI:10.1098/rspb.2004.2705 |
Cooke D E L, Drenth A, Duncan J M, et al. 2000. A molecular phylogeny of Phytophthora, and related oomycetes[J]. Fungal Genetics and Biology, 30(1): 17-32. DOI:10.1006/fgbi.2000.1202 |
Cooke D E L, Duncan J M. 1997. Phylogenetic analysis of Phytophthora species based on ITS1 and ITS2 sequences of the ribosomal RNA gene repeat[J]. Mycological Research, 101(6): 667-677. DOI:10.1017/S0953756296003218 |
Erwin D C, Ribeiro O K. 1996. Phytophthora Disease Worldwide. St. Paul, MN: American Phytopathological Society.
|
Forster H, Coffey M D. 1993. Molecular taxonomy of Phytophthora megasperma based on mitochondrial and nuclear DNA polymorphisms[J]. Mycological Research, 97(9): 1101-1112. DOI:10.1016/S0953-7562(09)80511-X |
Huai W X, Guo L D, He W. 2003. Genetic diversity of an ectomycorrhizal fungus tricholoma terreum in a larix principis-rupprechtii stand assessed using random amplified polymorphic DNA[J]. Mycorrhiza, 13(5): 265-270. DOI:10.1007/s00572-003-0227-8 |
Huai W X, Tian G, Hansen E M, et al. 2013. Identification of Phytophthora species baited and isolated from forest soil and streams in northwestern Yunnan Province, China[J]. Forest Pathology, 43(2): 87-103. DOI:10.1111/efp.2013.43.issue-2 |
Jung T, Chang T T, Bakonyi J, et al. 2017a. Diversity of Phytophthora species in natural ecosystems of Taiwan and association with disease symptoms[J]. Plant Pathology, 66(2): 194-211. DOI:10.1111/ppa.2017.66.issue-2 |
Jung T, Jung M H, Scanu B, et al. 2017b. Six new Phytophthora species from ITS clade 7a including two sexually functional heterothallic hybrid species detected in natural ecosystems in Taiwan[J]. Persoonia, 38: 100-135. DOI:10.3767/003158517X693615 |
Jung T, Nechwatal J. 2008. Phytophthora gallica sp.nov. a new species from rhizosphere soil of declining oak and reed stands in France and Germany[J]. Mycological Research, 112(10): 1195-1205. DOI:10.1016/j.mycres.2008.04.007 |
Jung T, Stukely M J C, Hardy G E S J, et al. 2011. Multiple new Phytophthora species from ITS clade 6 associated with natural ecosystems in Australia:evolutionary and ecological implications[J]. Persoonia Molecular Phylogeny and Evolution of Fungi, 26(69): 13-39. |
Katoh K, Standley D M. 2013. Mafft multiple sequence alignment software version 7:improvements in performance and usability[J]. Molecular Biology and Evolution, 30(4): 772-780. DOI:10.1093/molbev/mst010 |
Kroon L P N M, Bakker F T, van den Bosch G B M, et al. 2004. Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences[J]. Fungal Genetics and Biology, 41(8): 766-782. DOI:10.1016/j.fgb.2004.03.007 |
Kroon L P N M, Brouwer H, de Cock A W A M, et al. 2012. The genus Phytophthora anno 2012[J]. Phytopathology, 102(4): 348-364. DOI:10.1094/PHYTO-01-11-0025 |
Li W W, Zhao W X, Huai W X. 2017. Phytophthora pseudopolonica sp.nov. a new species recovered from stream water in subtropical forests of China[J]. International Journal of Systematic and Evolutionary Microbiology, 67(9): 3666-3675. DOI:10.1099/ijsem.0.002254 |
Martin F N, Abad Z G, Balci Y, et al. 2012. Identification and detection of Phytophthora: reviewing our progress, identifying our needs[J]. Plant Disease, 96(8): 1080-1103. DOI:10.1094/PDIS-12-11-1036-FE |
Martin F N, Blair J E, Coffey M D. 2014. A combined mitochondrial and nuclear multilocus phylogeny of the genus Phytophthora[J]. Fungal Genetics and Biology, 66(3): 19-32. |
Martin F N, Tooley P W. 2003. Phylogenetic relationships of Phytophthora ramorum, P. nemorosa, and P. pseudosyringae, three species recovered from areas in California with sudden oak death[J]. Mycological Research, 107(12): 1379-1391. DOI:10.1017/S0953756203008785 |
Martin F N, Tooley P W. 2004. Identification of Phytophthora isolates to species level using restriction fragment length polymorphism analysis of a polymerase chain reaction-amplified region of mitochondrial DNA[J]. Phytopathology, 94(9): 983-991. DOI:10.1094/PHYTO.2004.94.9.983 |
Nelson S C, Abad Z G. 2010. Phytophthora morindae, a new species causing black flag disease on noni (Morinda citrifolia L) in Hawaii[J]. Mycologia, 102(1): 122-134. DOI:10.3852/08-209 |
Reeser P W, Hansen E M, Sutton W. 2007. Phytophthora siskiyouensis, a new species from soil, water, myrtlewood (Umbellularia californica) and tanoak (Lithocarpus densiflorus) in southwestern Oregon[J]. Mycologia, 99(5): 639-643. DOI:10.1080/15572536.2007.11832528 |
Rizzo D M, Garbelotto M, Hansen E M. 2005. Phytophthora ramorum: integrative research and management of an emerging pathogen in California and Oregon forests[J]. Annual Review of Phytopathology, 43(1): 309-335. DOI:10.1146/annurev.phyto.42.040803.140418 |
Roy S G, Grünwald N J. 2014. The plant destroyer genus Phytophthora in the 21st century[J]. Review of Plant Pathology, 6: 387-412. |
Swofford D L. 2002. Paup*. Phylogenetic analysis using parsimony (*and other methods). version 4. 0b10. Mccarthy.
|
Uddin A J, Senda M, Uematsu S, et al. 2007. Phylogenetic relationship of Phytophthora citrophthora isolates based on rDNA internal transcribed spacer sequence analysis[J]. International Journal of Integrative Biology, 1(3): 150-156. |
Werres S, Marwitz R, Man I V W, et al. 2001. Phytophthora ramorum sp.nov. a new pathogen on Rhododendron and Viburnum[J]. Mycological Research, 105(10): 1155-1165. DOI:10.1016/S0953-7562(08)61986-3 |
White T J, Bruns T D, Lee S B, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols:A guide to methods and applications[M]. San Diego: Academic Press: 315-322.
|
Wills R T. 1992. The ecological impact of Phytophthora cinnamomi in the Stirling Range National Park, western Australia[J]. Austral Ecology, 18(2): 145-159. |
Xu P, Han Y, Wu J, et al. 2007. Phylogenetic analysis of the sequences of rDNA internal transcribed spacer (ITS) of Phytophthora sojae[J]. Acta Genetica Sinica, 34(2): 180-188. |
Yang X, Balci Y, Brazee N J, et al. 2016. A unique species in Phytophthora clade 10: Phytophthora intercalaris sp.nov. recovered from stream and irrigation water in the eastern USA[J]. International Journal of Systematic and Evolutionary Microbiology, 66(2): 845-855. DOI:10.1099/ijsem.0.000800 |
Zeng H C, Ho H H, Zheng F C. 2009. A survey of Phytophthora species on Hainan Island of south China[J]. Journal of Phytopathology, 157(1): 33-39. DOI:10.1111/jph.2008.157.issue-1 |
 2018, Vol. 54
2018, Vol. 54

