文章信息
- 孙化雨, 李利超, 赵韩生, 杨意宏, 王思宁, 高志民
- Sun Huayu, Li Lichao, Zhao Hansheng, Yang Yihong, Wang Sining, Gao Zhimin
- 黄藤2个NAC基因的分子特征及其SSR分子标记开发
- Molecular Characteristics and SSR Marker Development of Two NAC Genes from Daemonorops jenkinsiana
- 林业科学, 2017, 53(8): 132-140.
- Scientia Silvae Sinicae, 2017, 53(8): 132-140.
- DOI: 10.11707/j.1001-7488.20170815
-
文章历史
- 收稿日期:2016-09-12
- 修回日期:2016-10-24
-
作者相关文章
2. 河北农业大学园艺学院 保定 071001
2. College of Horticulture, Hebei Agricultural University Baoding 071001
NAC (NAM, ATAF和CUC)是植物所特有的转录因子家族,其成员数量众多。NAC家族成员的N端相对保守,都由约150个氨基酸组成结构域,且被分为5个亚结构域(A、B、C、D和E),但其C端的序列呈现出高度多样化,富含丝氨酸、谷氨酸等(Olsen et al., 2005)。迄今为止,对许多物种全基因组范围内的NAC转录因子进行了鉴定,包括拟南芥(Arabidopsis thaliana)(117个)、水稻(Oryza sativa)(151个)(Nuruzzaman et al., 2010)、大豆(Glycine max)(152个)(Le et al., 2011)、白菜(Brassica rapa)(204个)(Liu et al., 2014)、玉米(Zea mays)(152个)(Shiriga et al., 2014)、鹰嘴豆(Cicer arietinum)(71个)(Ha et al., 2014)、番茄(Solanum lycopersicum)(104个)(Su et al., 2015)和毛竹(Phyllostachys edulis)(125个)(黎帮勇等,2015)等。NAC转录因子的功能多样,参与许多生物学过程,包括花发育、次生壁形成、细胞分裂、顶端分生组织的形成、叶片衰老,以及生物和非生物胁迫应答(Olsen et al., 2005; Tran et al., 2010; Nakashima et al., 2012; Nuruzzaman et al., 2013; Banerjee et al., 2015)等,在植物的生长发育过程中起着重要的调控作用。
随着研究的不断深入,越来越多NAC在植物中的功能被揭示。NAM基因在矮牵牛(Petunia hybrida)的分生组织和原基边界表达,是其胚胎和花形成所需的(Souer et al., 1996);拟南芥CUC2的表达决定其叶缘锯齿的程度(Nikovics et al., 2006);玉米ZmCUC3在顶端分生组织外围表达,边界明显,部分细胞将形成一个新的叶原基(Zimmermann et al., 2005)。拟南芥NAC转录因子NTL8参与赤霉酸介导的盐信号调控(Kim et al., 2008),而且通过FLOWERING LOCUST调节拟南芥盐响应开花(Kim et al., 2007),冷激活NAC转录因子调控诱导拟南芥病原体的抗性反应(Seo et al., 2010),AtNAC2作为乙烯和生长素信号转导通路下游的转录因子,参与盐应激反应和侧根发育调控(He et al., 2005)。拟南芥NAC转录因子AtNAP在叶片衰老中发挥着重要作用,AtNAP的诱导表达导致拟南芥早熟衰老(Guo et al., 2006),ABA-AtNAP通过控制气孔的开闭来调节乙烯刺激呼吸,实现对果实的衰老调控(Kou et al., 2012)。在祖野麦(Triticum turgidum ssp. dicoccoides)中NAC转录因子(NAM-B1) 可以加速衰老,提高叶片的营养转运形成籽粒(Uauy et al., 2006)。水稻OsNAC19是一种转录激活因子,参与水稻响应稻瘟病菌感染,可能在茉莉酸甲酯介导的信号传导途径中起重要作用(Lin et al., 2007)。欧洲油菜(Brassica napus)BnaNAC55通过激活一些活性氧(Reactive Oxygen Species, ROS)以及与防御有关的基因的表达,来调控ROS的积累和细胞死亡(Niu et al., 2016)。
棕榈藤属于棕榈科(Palmae)植物,是重要的热带森林资源,其藤茎是优良的非木材森林产品之一,是编制各种高档家具及工艺品的理想材料,具有很高的经济价值。由于棕榈藤资源极其匮乏,其原料是国内外市场上的紧缺物资。绝大多数棕榈藤具有极强的攀缘性,总是和其他的树木纠缠在一起,使其成为森林的伴生物种。黄藤(Daemonorops jenkinsiana)为棕榈科鳞果亚科(Lepidocaryoideae)省藤族(CALAMEAE)黄藤属(Daemonorops)植物,分布于广东东南部、香港、海南及广西西南部,为我国棕榈科单属单种植物,叶羽状全裂,茎初时直立,后攀援,其攀援主要借助于叶片顶端延伸而成的具爪状刺的纤鞭。研究表明NAC参与叶片形态建成的调控(Zimmermann et al., 2005; Nikovics et al., 2006),目前虽然对黄藤的生长特性以及形态特征具有比较详细的描述(江泽慧等,2013),但关于黄藤的生长发育的分子基础尚属空白。因此,本研究以黄藤为对象,从叶片中分离克隆了2个NAC基因,并对其分子特征进行深入分析,研究其表达模式,并开发SSR分子标记,以期为深入研究其功能和分子标记辅助育种提供参考。
1 材料与方法 1.1 试验材料黄藤样品取自中国林业科学研究院热带林业研究所(广州,龙洞),分别采集叶片、发育初期的纤鞭、发育成熟的纤鞭、发育初期的钩刺、发育成熟的钩刺,放入RNAlater® RNA Stabilization Reagent(QIAGEN, 德国)中,带回实验室备用。另外从中国科学院西双版纳植物园分别采集的20个藤种(表 1)叶片,用硅胶干燥处理后带回实验室备用。
|
|
采用改良CTAB法提取不同藤种的基因组DNA(高志民等,2006),存-20 ℃备用。采用改良Trizol法提取黄藤叶片、发育初期钩刺、发育成熟钩刺、发育初期纤鞭和发育成熟纤鞭的总RNA(Gao et al., 2006),按照反转录试剂盒(Promega, 美国)说明书分别合成cDNA。
1.3 基因克隆与分析根据黄藤转录组数据(NCBI SRA注册号:SRR3089417、SRR3089429、SRR3089432、SRR3089433、SRR3089434、SRR3089435、SRR3089436和SRR3089437) 中预测的NAC转录因子基因序列设计引物,Teng-NAC3-F和Teng-NAC3-R,以及Teng-NAC4-F和Teng-NAC4-R(表 2),由生工生物工程股份有限公司合成。分别以黄藤cDNA和基因组DNA为模板进行扩增。PCR反应体系为:5×Prime STARTM Buffer 4 μL,dNTP Mixture (2.5 mmol·L-1)2 μL,正向引物(10 μmol·L-1)0.5 μL,反向引物(10 μmol·L-1)0.5 μL,cDNA/基因组DNA 2 μL,Prime STAR HS DNA polymerase 0.2 μL,DMSO 1 μL,加水补至总体积为20 μL。PCR反应程序为:98 ℃ 4 min;98 ℃ 30 s,60 ℃ 30 s,72 ℃ 1 min 50 s(基因组DNA扩增延伸为2 min 30 s),反应循环35个;72 ℃ 10 min,4 ℃保存。用胶回收试剂盒(Biomega, 美国)回收PCR产物,进行加A反应,反应体系为:10×LA PCR Buffer 1.0 μL,dATP(2.5 mmol·L-1)1.0 μL,回收产物7.9 μL,LA Taq酶0.1 μL;反应程序:70 ℃ 30 min。用加A产物连接pGEM-T easy载体(Promega, 美国),转化大肠杆菌(Escherichia coli)DH5α,选取阳性克隆,酶切检测分析后,送生工生物工程股份有限公司测序。
|
|
用DNAstar对测序获得的序列进行初步分析。用NCBI在线软件BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi)程序进行序列同源性比较。蛋白结构域预测用TMHMM 2.0 (http://myhits.isb-sib.ch/cgi-bin/motif_scan)来完成。对获得基因编码的蛋白采用Phyre2.0 (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index)进行三级结构建模。
1.4 基因表达分析分别利用定量引物qNAC3-F和qNAC3-R、qNAC4-F和qNAC4-R(表 2)对2个NAC基因在叶片、发育初期的纤鞭、发育成熟的纤鞭、发育初期的钩刺、发育成熟的钩刺中的表达变化情况进行定量分析。反应在耶拿qTOWER2.2仪器上进行,总体系(10.0 μL):5.0 μL LightCycler®480 SYBR Green I Master Mix (Roche, 美国),0.8 μL cDNA,正、反向引物各0.2 μL,3.8 μL ddH2O。反应程序:95 ℃ 5 min;95 ℃ 30 s,60 ℃ 10 s,40个循环。定量分析所用内参基因为γ-tubulin (GenBank No. KX793703)。反应重复3次,结果用2-ΔΔCt法(Livak et al., 2001)分析。
1.5 SSR位点分析利用SSRIT(Simple Sequence Repeat Identification Tool)在线软件(Temnykh et al., 2001)对获得的黄藤NAC转录因子基因序列进行SSR搜索,SSR类型最多核苷酸数量设置为4个,重复次数等于或大于5。
根据DjNAC3和DjNAC4基因组序列分别设计SSR引物(ssrNAC3-F和ssrNAC3-R、ssrNAC4-F和ssrNAC4-R),分别以21个不同藤种(表 1)的基因组DNA为模板,进行扩增,PCR产物用8%的PAGE电泳进行分析。回收以伸长钩叶藤、老挝钩叶藤、狭叶黄藤、黄藤属种1、勐腊鞭藤、斑岭省藤6个藤种基因组DNA为模板扩增获得的目的片段,并连接到pGEM-T easy载体,转化大肠杆菌,并送公司测序。
2 结果与分析 2.1 DjNAC的克隆与基因结构分析对应黄藤cDNA和基因组DNA模板,引物Teng-NAC3-F和Teng-NAC3-R的PCR扩增产物电泳结果显示,分别约在0.7 kb和0.8 kb有1条目的条带,同时引物Teng-NAC4-F和Teng-NAC4-R分别约在1.3 kb和1.4 kb有1条目的条带。分别回收目的条带测序结果表明,引物Teng-NAC3-F和Teng-NAC3-R扩增的cDNA为731 bp,包含1个729 bp的开放阅读(ORF),ORF对应的基因组序列为850 bp,包含2个外显子和1个121 bp内含子(454-574 bp),将该基因cDNA命名为DjNAC3 (GenBank登录号:KU556738)。引物Teng-NAC4-F和Teng-NAC4-R扩增的cDNA为1 334 bp,包含1个1 326 bp的ORF,ORF对应的基因组序列为1 441 bp,包含2个外显子和1个107 bp的内含子(875-981 bp),将该基因cDNA命名为DjNAC4 (GenBank登录号:KX579750)。DjNAC3和DjNAC4的开放阅读(ORF)对应的基因组所包含的内含子均符合GT-AG剪切原则(Moore et al., 1993),基因结构如图 1所示。

|
图 1 DjNAC3和DjNAC4的基因结构 Fig.1 Gene structures of DjNAC3 and DjNAC4 E:外显子;I:内含子。E: Exon; I: Intron. |
DjNAC3编码1个242 aa的蛋白,预测的分子量为27.95 kDa,等电点为8.323。DjNAC4编码1个441 aa的蛋白,预测的分子量为49.66 kDa,等电点为7.082。Blast分析表明,DjNAC3和DjNAC4具有典型的NAC转录因子结构特征,N端保守性较强,由大约150个高度保守的氨基酸残基组成,包含A、B、C、D、E 5个亚结构域,C端的氨基酸序列保守性较弱。DjNAC3和DjNAC4均属于CUC (development-related NAC)亚家族(Zhu et al., 2015),但二者之间的相似系数仅为23.6%,以c3ulxA_为模板构建DjNAC3和DjNAC4的三级结构(图 2),结构上的差异意味着它们在黄藤生长发育过程中可能具有不同的功能。
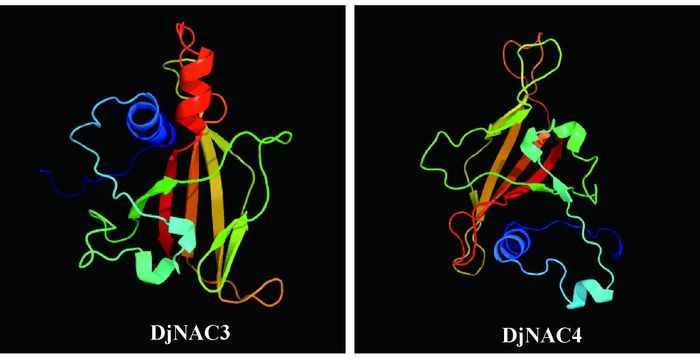
|
图 2 DjNAC3和DjNAC4蛋白的三级结构 Fig.2 Tertiary structures of DjNAC3 and DjNAC4 |
以黄藤叶片、发育初期钩刺、发育成熟钩刺、发育初期纤鞭和发育成熟纤鞭的cDNA为模板,进行实时定量PCR分析,检测DjNAC在不同组织中的表达模式。结果显示,DjNAC3和DjNAC4的表达模式存在着明显的差异。DjNAC3在叶片中的表达丰度最低,远低于钩刺和纤鞭,而纤鞭中的表达又高于钩刺,以发育成熟的纤鞭中的表达丰度最高;DjNAC4则在发育成熟的钩刺中表达丰度最高,发育成熟的纤鞭次之,叶片和发育初期的纤鞭中均较多,而发育初期的钩刺中最低(图 3)。由此表明,DjNAC3和DjNAC4在黄藤叶片发育过程中可能具有不同的功能。
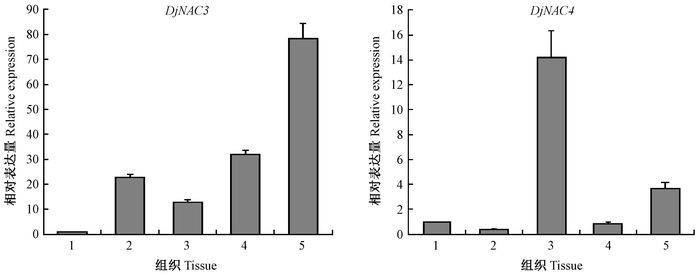
|
图 3 DjNAC3和DjNAC4在黄藤不同组织中的表达分析 Fig.3 Expression analysis of DjNAC3 and DjNAC4 in different tissues of Daemonorops jenkinsiana 1:叶片;2:发育初期钩刺;3:发育成熟钩刺;4:发育初期纤鞭;5:发育成熟纤鞭。 1: Leaves; 2: Early developed barbs; 3: Developed barbs; 4: Early developed cirri; 5: Developed cirri. |
SSR搜索结果表明,在DjNAC3和DjNAC4的基因组序列中分别找到1个SSR位点,其中前者的SSR位点位于内含子区域,SSR类型为TA,重复次数为6;后者的SSR位点位于第1个外显子区域,SSR类型为GCA,重复次数为5(表 3)。
|
|
为进一步验证SSR位点的可靠性,分别回收了伸长钩叶藤、老挝钩叶藤、狭叶黄藤、黄藤属种1、勐腊鞭藤、斑岭省藤6个藤种的PCR产物进行测序。结果表明,引物对ssrNAC3-F和ssrNAC3-R的扩增产物在205~220 bp之间,不同藤种样品中的SSR序列存在着一定的差异,既包括SSR类型变异、重复次数变化等多态性,又有SSR位点缺失的现象。如狭叶黄藤、黄藤属种1与黄藤的SSR位点均为(TA)6,而伸长钩叶藤、老挝钩叶藤和勐腊鞭藤的SSR位点均为(TA)4,少了2个TA重复,但在斑岭省藤中SSR位点却缺失了,同时在SSR位点之后的30个碱基各种之间存在着不同程度的多态性(图 4A)。引物对ssrNAC4-F和ssrNAC4-R的扩增产物在331~346 bp之间,其中狭叶黄藤、黄藤属种1与黄藤的SSR位点均为(GCA)5,而伸长钩叶藤、老挝钩叶藤、勐腊鞭藤和斑岭省藤的SSR位点分别为(GCA)8、(GCA)9、(GCA)7和(GCA)4,不同属间存在明显的多态性(图 4B)。

|
图 4 不同藤种样品SSR扩增产物的测序分析 Fig.4 Analysis of SSR sequences of PCR products amplified from six rattan samples A:ssrNAC3-F和ssrNAC3-R扩增产物序列;B:ssrNAC4-F和ssrNAC4-R扩增产物序列。编号与表 1中藤种相对应。 A: Sequences of products amplified by ssrNAC3-F and ssrNAC3-R; B: Sequences of products amplified by ssrNAC4-F and ssrNAC4-R. The No. represents the species in Tab. 1 correspondingly. |
为验证SSR位点的通用性和多态性,利用SSR引物(ssrNAC3-F和ssrNAC3-R、ssrNAC4-F和ssrNAC4-R)(表 2),以黄藤和另外20个不同藤种的基因组DNA为模板进行扩增。PAGE电泳结果表明,所有PCR产物均有明显的目的条带,表明2个分子标记在不同藤种中具有通用性(图 5)。

|
图 5 不同藤种样品SSR引物扩增产物的PAGE电泳分析 Fig.5 PAGE analysis of PCR products amplified from different rattan samples using SSR primers A:ssrNAC3-F和ssrNAC3-R的扩增产物;B:ssrNAC4-F和ssrNAC4-R的扩增产物。M:DNA分子量标记;1-21:不同藤种样品。 A. PCR products of ssrNAC3-F and ssrNAC3-R; B. PCR products of ssrNAC4-F and ssrNAC4-R. M: DNA ladder; 1-21: Different rattan samples. |
引物对ssrNAC3-F和ssrNAC3-R的21个样品扩增产物约在220 bp均有一明显特异条带(图 5A),其中钩叶藤属的伸长钩叶藤和老挝钩叶藤的目的条带大小一致,而细钩叶藤的明显偏大;黄藤属的长柄黄藤、狭叶黄藤和黄藤种1的带型一致,而黄藤的目的条带明显偏小;省藤属中除了褐鳞省藤和勐捧省藤带型一致、种2(C. sp. 2) 和种3(C. sp. 3) 的带型较一致外,其他藤种的带型差异明显。引物对ssrNAC4-F和ssrNAC4-R的扩增产物则约在350 bp均有一明显特异条带(图 5 B),其中钩叶藤属3个藤种的目的条带大小一致;黄藤属的长柄黄藤、狭叶黄藤和种1(D. sp. 1) 的带型一致,而黄藤的目的条带明显偏小;省藤属柳条省藤和褐鳞省藤带型目的条带差异明显,而褐鳞省藤和勐捧省藤的相一致,其余藤种的带型差异明显,但元江省藤(C. sp.)与种1(C. sp. 1)、种2( C. sp. 2) 和种3(C. sp. 3) 的基本一致。由此表明,2个SSR分子标记在不同藤种中具有多态性,尤其是不同属的不同藤种之间更为明显。
3 讨论NAC转录因子对植物不同组织器官的系统发育具有重要作用,是植物形态建成中不可或缺的调控因子。根据蛋白结构特征将被子植物的NAC划分为6个大的家族,每个家族包含不同的亚家族,即家族Ⅰ(VND和NST/BRN/SMB)、Ⅱ(NAC1、CUC和ORE)、Ⅲ(NTL、NAC2和TMM)、Ⅳ(FEZ/JUB和LOV1)、Ⅴ(XND1、胁迫相关家族和NARS)和家族Ⅵ(其他) (Pereira-Santana et al., 2015)。本研究从黄藤叶片中分离得到的DjNAC3和DjNAC4基因均属于NAC家族的CUC亚家族。基因结构的差异,意味着它们具有不同的功能,DjNAC3和DjNAC4在叶片、发育初期钩刺、发育成熟钩刺、发育初期纤鞭和发育成熟纤鞭中的基因表达模式差异进一步证实了这一点。纤鞭是棕榈藤植物为适应森林攀援生活形成的最为重要的器官,是由叶轴顶端延伸出去形成的,或是叶鞘发育形成。黄藤的叶片属于大型羽状复叶(全裂),其纤鞭由叶轴顶端延伸发育而成,纤鞭上具有轮生的钩刺,利于绞缠攀爬。DjNAC3和DjNAC4在黄藤叶片的不同部位均有表达,且存在明显差异,但其功能如何,在黄藤叶片生长发育中是怎么实现其调控作用的,有待于进一步深入研究。
SSR标记作为一种有效的分子标记已被广泛应用于遗传多样性分析、系统发育、辅助分类和育种研究等。DjNAC3和DjNAC4的基因组中均具有1个SSR位点,并证实在21个不同棕榈藤样品中具有通用性,且在不同属种间具有多态性,这为进一步研究棕榈藤的遗传多样性和辅助分类提供了分子标记。另外研究表明,SSR位点在基因序列中所处位置不同,其功能也存在着差异,如位于5′非翻译区的SSR可以调控基因的转录和翻译,位于基因内部的SSR具有较强的选择压力,可以调控基因的表达,位于内含子区域的SSR可以影响基因的转录,位于3′非翻译区的SSR可以引起mRNA的延伸(Li et al., 2004)。DjNAC3和DjNAC4的SSR位点,分别位于内含子区域和外显子区域,这对二者的基因表达可能具有不同的影响,具体情况需要进一步试验验证。
随着现代生物技术的快速发展,利用基因工程手段开发利用植物中具有调控作用的关键NAC转录因子,实现植物性状的定向改良已成为现实。水稻SNAC1转入小麦(Triticum aestivum)导致转基因植物生长迟缓(Saad et al., 2013),毛竹PeNAC在拟南芥中过量表达促进转基因植株的侧根发育(Wang et al., 2016),在水稻中过量表达穇子(Eleusine coracana)的EcNAC67能够提高水稻的耐盐和抗旱能力(Rahman et al., 2016),大麦(Hordeum vulgare)中过量表达HvNAC005导致提早成熟(Christiansen et al., 2016)。棕榈藤植物作为重要的森林资源之一,其潜在的价值远远没有得到开发利用,随着对其生长发育分子基础研究的不断深入,诸如NAC家族的转录因子等具有重大经济价值的功能基因将不断被挖掘出来,对其性状的调控将更具有操作性。因此,未来棕榈藤的分子育种有着广阔的前景。
4 结论本研究从黄藤中克隆了2个NAC转录因子家族成员基因DjNAC3和DjNAC4,二者在基因结构、编码蛋白特征等方面具有明显不同的分子特征,在黄藤叶片形态建成中可能具有不同的调控功能。同时,DjNAC3和DjNAC4的序列中均包含1个SSR位点,分别为(TA)6和(GCA)5,据此开发的分子标记在不同的棕榈藤中具有较高的通用性和多态性,可应用于棕榈藤辅助分类和分子辅助育种。
| [] |
高志民, 范少辉, 高健, 等. 2006. 基于CTAB法提取毛竹基因组DNA的探讨. 林业科学研究, 19(6): 725–728.
( Gao Z M, Fan S H, Gao J, et al. 2006. Extract genomic DNA from Phyllostachys edulis by CTAB-based method. Forest Research, 19(6): 725–728. [in Chinese] ) |
| [] |
江泽慧, 王康林. 2013. 中国棕榈藤. 北京, 科学出版社: 90-92.
( Jiang Z H, Wang K L. 2013. Rattan in China. Beijing, Science Press: 90-92. [in Chinese] ) |
| [] |
黎帮勇, 胡尚连, 曹颖, 等. 2015. 毛竹NAC转录因子家族生物信息学分析. 基因组学与应用生物学, 34(8): 1769–1777.
( Li B Y, Hu S L, Cao Y, et al. 2015. Bioinformatics analysis of NAC gene family in moso bamboo. Genomics Applied Biol, 34(8): 1769–1777. [in Chinese] ) |
| [] | Banerjee A, Roychoudhury A. 2015. WRKY proteins:signaling and regulation of expression during abiotic stress responses. Sci World J, 2015: 807560. |
| [] | Christiansen M W, Matthewman C, Podzimska-Sroka D, et al. 2016. Barley plants over-expressing the NAC transcription factor gene HvNAC005 show stunting and delay in development combined with early senescence. J Exp Bot, 67(17): 5259–5273. DOI:10.1093/jxb/erw286 |
| [] | Gao Z M, Li X P, Li L B, et al. 2006. An effective method for total RNA isolation from bamboo. Chinese For Sci Tech, 5(3): 52–54. |
| [] | Guo Y, Gan S. 2006. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J, 46(4): 601–612. DOI:10.1111/tpj.2006.46.issue-4 |
| [] | Ha C V, Esfahani M N, Watanabe Y, et al. 2014. Genome-wide identification and expression analysis of the CaNAC family members in chickpea during development, dehydration and ABA treatments. PLoS One, 9(12): e114107. DOI:10.1371/journal.pone.0114107 |
| [] | He X J, Mu R L, Cao W H, et al. 2005. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J, 44(6): 903–916. DOI:10.1111/tpj.2005.44.issue-6 |
| [] | Kim S G, Kim S Y, Park C M. 2007. A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUST in Arabidopsis. Planta, 6(3): 647–654. |
| [] | Kim S G, Lee A K, Yoon H K, et al. 2008. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J, 55(1): 77–88. DOI:10.1111/tpj.2008.55.issue-1 |
| [] | Kou X, Watkins C B, Gan S S. 2012. Arabidopsis AtNAP regulates fruit senescence. J Exp Bot, 63(17): 6139–6147. DOI:10.1093/jxb/ers266 |
| [] | Le D T, Nishiyama R, Watanabe Y, et al. 2011. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res, 18(4): 263–276. DOI:10.1093/dnares/dsr015 |
| [] | Li Y C, Korol A B, Fahima T, et al. 2004. Microsatellites within genes:structure, function, and evolution. Mol Biol Evol, 21(6): 991–1007. DOI:10.1093/molbev/msh073 |
| [] | Lin R M, Zhao W S, Meng X B, et al. 2007. Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Sci, 172(1): 120–130. DOI:10.1016/j.plantsci.2006.07.019 |
| [] | Liu T K, Song X M, Duan W K, et al. 2014. Genome-wide analysis and expression patterns of NAC transcription factor family under different developmental stages and abiotic stresses in Chinese cabbage. Plant Mol Biol Report, 32(5): 1041–1056. DOI:10.1007/s11105-014-0712-6 |
| [] | Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods, 25: 402–408. DOI:10.1006/meth.2001.1262 |
| [] | Moore M J, Query C C, Sharp P A. 1993. Splicing of precursors to mRNA by the spliceosome//Gesteland R F, Atkins J F. The RNA world. Cold Spring Harbor:Cold Spring Harbor Laboratory Press, 303-357. |
| [] | Nakashima K, Takasaki H, Mizoi J, et al. 2012. NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta, 1819(2): 97–103. DOI:10.1016/j.bbagrm.2011.10.005 |
| [] | Nikovics K, Blein T, Peaucelle A, et al. 2006. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell, 18(11): 2929–2945. DOI:10.1105/tpc.106.045617 |
| [] | Niu F, Wang C, Yan J, et al. 2016. Functional characterization of NAC55 transcription factor from oilseed rape (Brassica napus L.) as a novel transcriptional activator modulating reactive oxygen species accumulation and cell death. Plant Mol Biol, 92(1/2): 89–104. |
| [] | Nuruzzaman M, Manimekalai R, Sharoni A M, et al. 2010. Genome-wide analysis of NAC transcription factor family in rice. Gene, 465(1/2): 30–44. |
| [] | Nuruzzaman M, Sharoni A M, Kikuchi S. 2013. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol, 4: 248. |
| [] | Olsen A N, Ernst H A, Leggio L L, et al. 2005. NAC transcription factors:structurally distinct, functionally diverse. Trends Plant Sci, 10(2): 79–87. DOI:10.1016/j.tplants.2004.12.010 |
| [] | Pereira-Santana A, Alcaraz L D, Castaño E, et al. 2015. Comparative genomics of NAC transcriptional factors in angiosperms:Implications for the adaptation and diversification of flowering plants. PLoS One, 10(11): e0141866. DOI:10.1371/journal.pone.0141866 |
| [] | Rahman H, Ramanathan V, Nallathambi J, et al. 2016. Over-expression of a NAC 67 transcription factor from finger millet (Eleusine coracana L.) confers tolerance against salinity and drought stress in rice. BMC Biotechnol, Suppl 1: 35. |
| [] | Saad A S, Li X, Li H P, et al. 2013. A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses. Plant Sci, 203/204: 33–40. DOI:10.1016/j.plantsci.2012.12.016 |
| [] | Seo P J, Kim M J, Park J Y, et al. 2010. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J, 61(4): 661–671. DOI:10.1111/tpj.2010.61.issue-4 |
| [] | Shiriga K, Sharma R, Kumar K, et al. 2014. Genome-wide identification and expression pattern of drought-responsive members of the NAC family in maize. Meta Gene, 2: 407–417. DOI:10.1016/j.mgene.2014.05.001 |
| [] | Souer E, van Houwelingen A, Kloos D, et al. 1996. The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell, 85(2): 159–170. DOI:10.1016/S0092-8674(00)81093-4 |
| [] | Su H, Zhang S, Yin Y, et al. 2015. Genome-wide analysis of NAM-ATAF1, 2-CUC2 transcription factor family in Solanum lycopersicum. J Plant Biochem Biotechnol, 24(2): 176–183. DOI:10.1007/s13562-014-0255-9 |
| [] | Temnykh S, DeClerck G, Lukashova A, et al. 2001. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.):frequency, length variation, transposon associations, and genetic marker potential. Genome Res, 11(8): 1441–1452. DOI:10.1101/gr.184001 |
| [] | Tran L S, Nishiyama R, Yamaguchi-Shinozaki K, et al. 2010. Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops, 1(1): 32–39. DOI:10.4161/gmcr |
| [] | Uauy C, Distelfeld A, Fahima T, et al. 2006. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science, 314(5803): 1298–1301. DOI:10.1126/science.1133649 |
| [] | Wang L, Zhao H, Chen D, et al. 2016. Characterization and primary functional analysis of a bamboo NAC gene targeted by miR164b. Plant Cell Rep(6): 1371–1383. |
| [] | Zhu G, Chen G, Zhu J, et al. 2015. Molecular characterization and expression profiling of NAC transcription factors in Brachypodium distachyon L. PLoS One, 10(10): e0139794. DOI:10.1371/journal.pone.0139794 |
| [] | Zimmermann R, Werr W. 2005. Pattern formation in the monocot embryo as revealed by NAM and CUC3 orthologues from Zea mays L. Plant Mol Biol, 58(5): 669–685. DOI:10.1007/s11103-005-7702-x |
 2017, Vol. 53
2017, Vol. 53

