文章信息
- Cao Yabing, Zhai Xiaoqiao, Deng Minjie, Zhao Zhenli, Fan Guoqiang
- 曹亚兵, 翟晓巧, 邓敏捷, 赵振利, 范国强
- Relationship between Metabolites Variation and Paulownia Witches' Broom
- 泡桐丛枝病发生与代谢组变化的关系
- Scientia Silvae Sinicae, 2017, 53(6): 85-93.
- 林业科学, 2017, 53(6): 85-93.
- DOI: 10.11707/j.1001-7488.20170610
-
文章历史
- Received date: 2016-02-22
- Revised date: 2017-03-11
-
作者相关文章
2. Forestry Academy of Henan Zhengzhou 450008
2. 河南省林业科学研究院 郑州 450008
Paulownia tomentosa is a fast-growing, adaptable, country-yard, greening trees that native to China, where it has been cultivated for more than 2 000 years for its rot resistance, dimensional stability, desert control, high ignition point, and straight-grained timber, which made them widely used in house construction, papermaking, high-grade furniture, and handicrafts production (Bayliss et al., 2005; Bassett, 1998). To date, Paulownia trees have been introduced into many other countries (Ipekci et al., 2003; Ates et al., 2008). Paulownia witches' broom (PaWB), caused by phytoplasma, is a devastating disease of Paulownia trees that can result in significant economic losses in Paulownia plantation regions. Infected Paulownia trees are characterized by witches' brooms, short internodes, yellowing or reddening of leaves, phyllody, stunting and decline, virescence, sterile flowers and necrosis, and altered volatile production(Namba, 2002). In the past 40 years, researches on PaWB were mainly focused on the ecological and biological characteristics, the route of pathogen transmission, and the prevention of insect vectors during the phytoplasma infection (Jin et al., 1981; Cao et al., 2012; Fan et al., 2007). Recently, studies at the molecular level were also carried out, including transcriptom, miRNA, degradome and DNA methylation, which revealed some key genes and miRNAs that related to PaWB (Liu et al., 2013; Mou et al., 2013; Fan et al., 2014; Cao et al., 2014a; 2014b; Niu et al., 2016). However, the molecular mechanisms associated with the tree's response to phytoplasma infection are still largely unknown. As we know, metabolites are the end products of gene expression, to some extent, which may be more accurate than genes in response to interactions between plants and pathogens (Ward et al., 2007). Therefore, it is necessary to carried out metabonomics research to investigate changes of metabolites in Paulownia with phytoplasma.
Metabonomics is a term that used to describe qualitative and quantitative analysis of the small molecule metabolites (relative molecular weight < 1 000 dalton) in organism, cell, or tissue, The term is used widely in system biology(Allwood et al., 2008; Wilson et al., 2005). Metabonomic analysis can help reveal the complex interaction of metabolic compositions and play important roles in biotic and abiotic stress, which usually represented by abnormal changes in metabolites in tissues (Ward et al., 2014). Metabolites related to salt stress and phytoplasma infection in Synechocystis sp. and grapevine have been reported using metabonomics analysis (Gai et al., 2014; Wang et al., 2014). In recent years, a series of secondary metabolites in different organs of P. tomentosa have been isolated and identified(Schneiderová et al., 2015). But there have few reports on metabonomic changes in P. tomentosa with PaWB until now. Here, we aimed to detect metabolite changes with PaWB and to identify metabolites that showed differential expression between the healthy and the phytoplasma-infected P. tomentosa seedlings. To our knowledge, this is the first report on metabolite variations in P. tomentosa with witches' broom. This result provides a comprehensive foundation for us to understand the mechanism of PaWB.
1 Materials and methods 1.1 PlantTissue-cultured seedlings of P. tomentosa were obtained from the Forest Biotechnology Laboratory of Henan Agricultural University, Zhengzhou, Henan Province, China. After all the seedlings were cultured in 100 mL triangular flasks on 1/2 Murashige and Skoog (MS) medium which did not contain any phytohormone for 30 days, the uniformly grown terminal buds from the healthy seedlings (HS) and phytoplasma-infected seedlings (PIS) were transferred into 100 mL triangular flasks containing 1/2 MS culture medium supplemented with 25 mg·L-1 sucrose and 8 mg·L-1 agar (Sangon, Shanghai, China), and cultured under the same conditions with air temperature maintained at 25 ± 2 ℃ and 130 μmol·m-2 s-1 intensity light (with 16/8 h light/dark photoperiod). After 30 days, terminal buds about 1.5 cm in length were collected from 60 healthy seedlings (or phytoplasma-infected seedlings), then frozen in liquid nitrogen and stored at -80 ℃ for later analysis.
1.2 Detection of PaWB phytoplasma in P. tomentosaTotal DNA was extracted from the terminal buds of HS and PIS, respectively, according to the method as described by Zhang et al., (2009). The DNAs were assessed with NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA). PaWB phytoplasma was detected by nested-PCR as described by Lee et al., (1993). The PCR procedure and agarose gel electrophoresis were performed according to the method as described by Fan et al., (2007).
1.3 Metabolite extraction and analysis by high performance liquid chromatography-mass spectrometry (HPLC-MS)The HS and PIS terminal buds (50 mg) stored at -80 ℃ were ground in a mortal using liquid nitrogen, respectively, and then extracted with 1 mL of 50% methanol (pre-cooled at -20 ℃). Subsequently, the samples were vortexed for 30 s and precipitated for 20 min. Following centrifugation (10 000 r·min-1, 10 min, 4 ℃), 250 μL of supernatant was transferred into 1.5 mL polypropylene tubes, and 10 μL of the supernatant was injected into a HPLC-MS system. A 'quality control' (QC) sample was also prepared by mixing tissue powder, then following the metabolite extraction step as described above.
The analytical HPLC-MS data were evaluated using shimadzu HPLC system (shimadzu, Japan) coupled to a LTQ Orbitrap Velos instrument (Thermo Fisher Scientific, MA, USA) set at 70 000 resolutions, and samples analysis were carried out in the positive ion modes. The scanning range of the mass was 50-1 500 m/z. The flow rate of nitrogen sheath gas was set at 30 L·min-1 and the nitrogen auxiliary gas at 10 L·min-1. Spray volage was set to 4.5 kV. The mobile phase consisted of solvent A (5% V/V formic acid/water), and solvent B (100% acetonitrile). The flow rate was 0.2 mL·min-1 and the injection volume was 10 μL. An Agilent Zorbax ODS C-18 column (Agilent Technologies, Santa Clara, CA, 150 mm×2.1 mm, 3.5 μm) was used for all analysis. The gradient that we used was similar to that used for the serum metabolomics analysis(Luan et al., 2013) with some modifications: 5% solvent B at 0-2 min, 7% B at 2-3 min, 8% B at 3-4 min, 70% B at 4-5 min, 90% B at 5-9 min, 95% B at 9-11 min, 95%-5% B at 11-14 min and 5% B at 14-15 min. To ensure the system balance, the pooled "QC" sample was injected five times at the beginning of the run, and then each 10 samples to further monitor the stability of the analysis (Want et al., 2010).
1.4 Data analysisPretreatment of the data including peak picking, peak grouping, retention time correction and second peak grouping was obtained through the XCMS software implemented with the freely available R statistical language (v2.13.1). The initially mass spectrum data cannot be directly used in the analysis; therefore, the HPLC-MS raw data files were firstly converted into XML format and then directly processed by the XCMS toolbox (http://metlin.scripps.edu/xcms). A list of the ion intensities of each peak detected was generated using retention time and the m/z data pairs as identifiers for each ion. Finally, a three-dimensional matrix was obtained, which contained arbitrarily assigned peak indices, retention times-m/z pairs, and ion intensity information (variables).
External factors can affect the reliability of the subsequent analysis, therefore, to obtain consistent variables, we implemented a series of measures such as removing peaks with more than 80% missing values (those with ion intensity=0) and those geometric isomer, which further reduced the resulting matrix.To ensure data were of comparable high quality within an analytical run, a method based on the periodic analysis of "QC" sample together with the PIS and HS samples is used as a quality assurance strategy in our present study. Here each retained peak is normalized to the QC sample using robust Loess signal correction (R-LSC). A threshold of 30% was set for the relative standard deviation value of metabolites in the "QC"samples, which is accepted as a standard in the assessment of repeatability in metabolomics datasets. Finally, the filtered matrix was exported for multivariate statistical analysis using principal component analysis (PCA) and partial least-squares discriminant analysis (PLS-DA).
Each metabolite peak reproducibly detected in whole samples was measured using the univariate method Welch's t Test to estimate the significance of each metabolite among the two samples, with the results adjusted for multiple testing using false discovery rate (FDR) correction.On the bases of a variable importance in the projection (VIP) threshold of 1 from the 7-fold cross-validated PLS-DA model, a number of metabolite ions responsible for the difference in the metabolic profile scan of HS and PIS were obtained. The metabolite ions identified by the PLS-DA model were validated at a univariate level using the FDR test from the R statistical toolbox with the critical P value set to < 0.05. The corresponding fold changes (fold change > 1.5, or < 0.67) show how these selected differentially expressed metabolites varied between the two groups (HS and PIS). Discriminatory metabolites of interest were extracted from the combining VIP value, P value and fold change.
Exact molecular mass data from redundant m/z peaks were used for metabolite searches against the KEGG database. A metabolite name was reported when a match with a mass difference between observed and theoretical mass was < 20×10-6. The isotopic distribution measurement was used to further validate the metabolite molecular formula of matched metabolites.
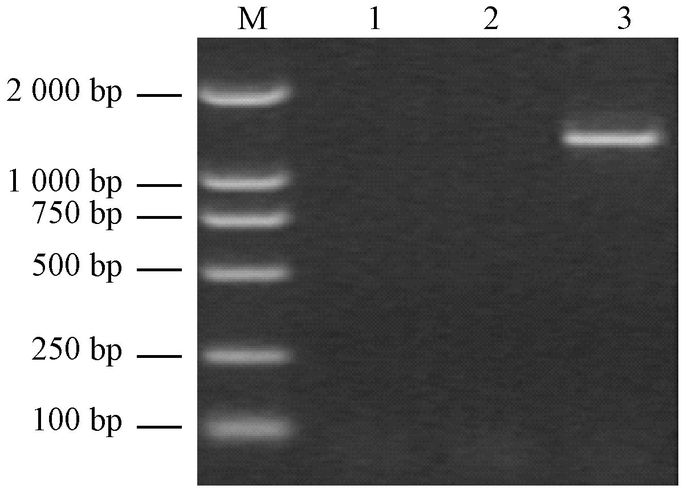
2 Results 2.1 Detection of PaWB phytoplasma in the Paulownia seedlingsThe PIS were characterizedwith witches' broom, yellow of leaves, short internodes and proliferation of axillary buds, while the HS don't showed none of these morphological symptoms (Fig. 1). Based on the PaWB phytoplasma genome, we used a nested-PCR to amplify its 16Sr DNA fragments. The specific 1.2 kb fragments of phytoplasma genome was detected only in the PIS (Fig. 2), indicating that the two samples were satisfied for the metabolomic analysis.

|
Fig.1 Changes of morphology in Paulownia witches' broom seedlings A: Phytoplasma-infected seedlings; B: Healthy seedlings. |
|
Fig.2 Detection of phytoplasma 16S rRNA in phytoplasma-infected seedlings M: Marker; 1: Healthy seedlings; 2: ddH2O; 3: Phytoplasma-infected seedlings. |
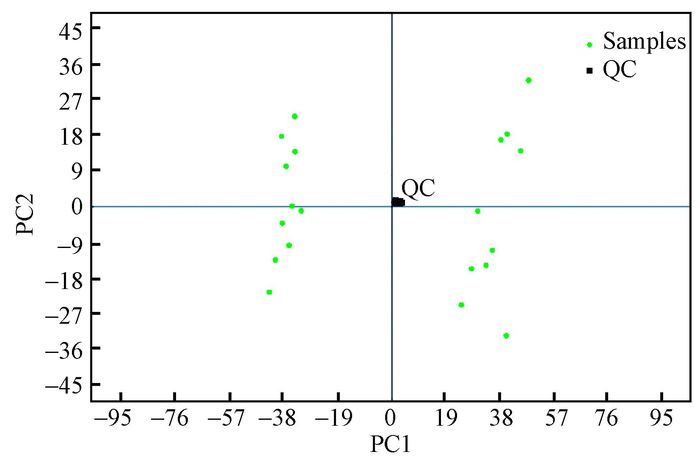
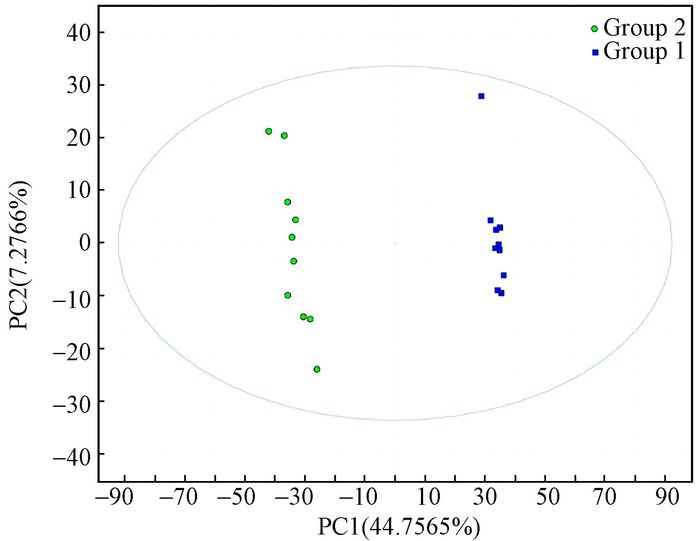
After running the optimized HPLC-MS analysis protocol and subsequent data processes, 1 466 retention time-exact mass pairs remaining in each sample profile and analyzed under positive ion mode. PCA showed that no shift was observed in the space of data obtained from the 'QC' samples (Fig. 3). This result implied that extracted and detected metabolites from the PIS and the HS with fairly consistent retention times and excellent resolution in the HPLC-MS analysis. All the variables were used in the following analysis. Significant differences in metabolic profile between the HS and PIS were also observed using a two components PCA score plot. Then, the cross-validated two-component PLS-DA demonstrated satisfactory modeling and achieved a distinct separation between metabolite profiles of the HS and PIS as shown in Fig. 4. Fig. 4 showed the PLS-DA score plot. From the corresponding loading plots and with the VIP≥1, 766 ions furthest away from the origin were found significantly contribute to the separation and, therefore, may be regarded as the differential m/z. At the same time, 1 271 metabolite m/z that displayed large-magnitude-changes were also detected by a volcano plot and a statistical test (P-value < 0.05, fold change > 1.5, or < 0.67). Of these peaks, 571 were ultimately selected as discriminating m/z through a combined analysis of their VIP values and volcano plot results. These metabolites may play important roles in phytoplasma infection.
|
Fig.3 PCA plot for consecutively analyzed QC samples |
|
Fig.4 Scores plot of the PLS-DA model for discriminating healthy seedlings from phytoplasma-infected seedlings Group1: Healthy seedlings; Group2: Phytoplasma-infected seedlings. |
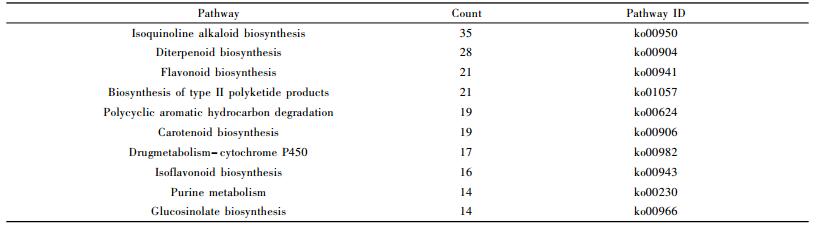
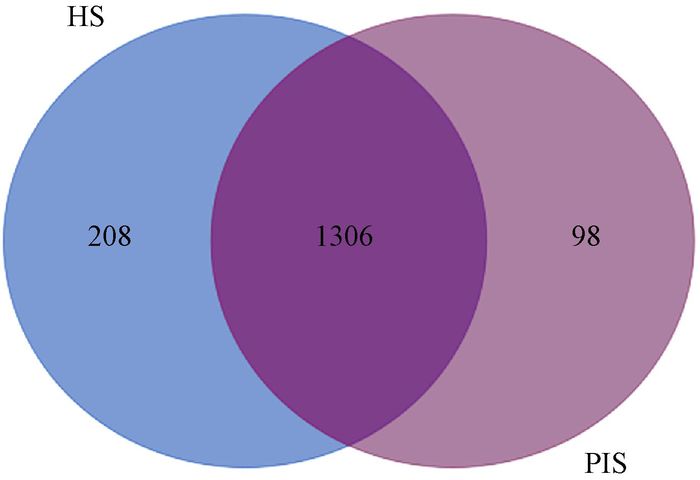
To identify the possible metabolites related to phytoplasma infection, all the 571 differentially expressed peaks were first aligned to KEGG metabolites database with a mass difference between observed and theoretical mass < 20×10-6. A total of 373 peaks were matched 1 612 known metabolites, while 198 did not match any known metabolites. To identify metabolites related to PaWB, the expression levels of the 1 612 differentially expressed metabolites in HS vs. PIS were investigated, among them, 208 only existed in HS, 98 only existed in PIS, and 1 306 existed in both of them(Fig. 5). By further analysis of these 1 612 differentially expressed metabolites, 765 had decreased expression levels (fold-change < 0.67) in PIS. Compared to HS, and 847 metabolites are on the contrary (fold-change > 1.5). Based on the KEGG annotation, 460 differentially expressed metabolites were mapped to 111 KEGG pathways (Tab. 1). The most significantly represent categories among the assigned pathways were "Isoquinoline alkaloid biosynthesis"(Ko00950, 35, 19 increased and 16 decreased), followed by "Diterpenoid biosynthesis"(Ko00904, 28, 8 increased and 20 decreased)" and "Flavonoid biosynthesis"(Ko00941, 21, 15 increased and 6 decreased).
|
Fig.5 Venn plot for 1 612 differentially expressed metabolites HS: Healthy seedlings; PIS: Phytoplasma-infected seedlings. |
|
|
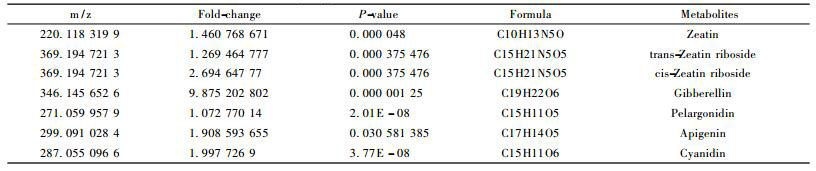
When faced with phytoplasma invasion, the host plant activates a set of physiology and biochemistry response like changes inthe concentration or types of metabolites. In our study, some metabolites which cannot be detected as genes encode for them were inhibited in phytoplasma-infected seedlings, for instance, niguldipine and berbamunine; while some metabolites were on the contrary, such as cupreine and all-trans-hexaprenyl diphosphate; besides, there were others metabolite that present in both healthy and phytoplasma-infected seedlings, but showed significant difference, including phenylalanine, phyllanthin, clitidine 5′-phosphate, zeatin, anthocyanin and so on. To further validate the metabolites that closely related to PaWB, functional analysis of these metabolites were investigated, and some differentially expressed metabolites related to PaWB were found out as shown in Tab. 2. Among them, the most significant one was gibberellin, followed by zeatin riboside, cyanidin, apigenin and pelargonidin. KEGG pathway analysis showed that these differentially expressed metabolites were mainly involved in zeatin biosynthesis, flavonoid biosynthesis, and plant hormone signal transduction, indicating that phytoplasma infection activated the defense reaction, including the burst of active oxygen and callose deposition, and disturbed the plant metabolic process, triggering the hormonal imbalances in Paulownia. All these effects in turn enhanced the concentration of antioxidant and phytohormones, which possibly suppress the defense systems and promote the formation of the typical symptoms in Paulownia.
|
|
In this study, we analyzed changes in P. tomentosa metabolites in response to PaWB phytoplasma infection. We obtained a set of 1 612 metabolites, among which changes of hormones and anthocyanin metabolism may be closely related to PaWB.
3.1 Approaches used to detect metabolites in organisms, cells, and tissuesMetabonomics aims to describe thenon-targeted "global" analysis of tissues and biofluids for low molecular mass organic endogenous metabolites regardless of whether their chemical structures are known(Krall et al., 2009). Therefore, in view of the advantages to detect subtle differences in the complex mixtures, metabonomics has been used in plants to found metabolites that related to disease. Over the past decade, several methods that offer high accuracy and sensitivity for analysis of highly complex mixtures of compounds have been established, including nuclear magnetic resonance (NMR), gas chromatography-mass spectrometry (GC-MS), mass spectrometry (MS), liquid chromatography-mass spectrometry (LC-MS), high performance liquid chromatography (HPLC). Among these approaches, NMR has the advantages on high information concentration of the spectra and the relative stability of NMR-chemical shifts(Wilson et al., 2005), but the spectras contain many overlapping peaks and have a low sensitivity(Ward et al., 2007); though GC-MS has a high reproducibility and a relatively broad coverage of compound classes, it not sufficient for analysis of some plant metabolites that are volatile or that can pass the GC separation under the conditions applied (Roessner et al., 2000). While the HPLC-MS has the potential to generate more metabolite profiles and can provide a valuable complementary information and molecule structure of metabolites compared to other methods. Today, HPLC-MS has been used to analyze the thermal instability or low concentration compounds (Wilson et al., 2005; Krall et al., 2009). Therefore, we used HPLC-MS in this study to analyze the metabolites of P. tomentosa.
3.2 Anthocyanin metabolism responding to phytoplasma infection in P. tomentosaDuring pathogen infection, a series of complex biological and molecular mechanism may be induced to resist to the pathogens in plants, including accumulation of reactive oxygen species and expression of pathogenesis-related proteins (Zipfel et al., 2010; Torres et al., 2006). Previous study has demonstrated that antioxidant plays important role in the defense systems of Paulownia in response to phytoplasma infection like p450 (Liu et al., 2013). In our present study, we indentified similarly functional metabolites. Anthocyanins, a major class of secondary metabolites in plant, are synthesized in the flavonoid pathway and act as antioxidant in biotic and abiotic stress, which can prevent the plant cells from damage that caused by reactive oxygen species (ROS) (Gould et al., 2002; Margaria et al., 2014; Berli et al., 2010). However, high level of anthocyanins may reduce the defense reactions triggered by the host plant and lead to an increase of phytoplasma growth. In pathogens-infected plants, callose deposition can inhibit the transport of carbohydrate through sieve tubes and cause the structure damage of chloroplast, leading to increased accumulation of ROS (Ji et al., 2009; Durrant et al., 2000). ROS, a normal characteristic event of pathogen infection plants, usually acts as secondary messengers and plays a central role in the defense of plants against pathogens (Gai et al., 2014). To some extent, excess H2O2 accumulation can lead to oxidative stress, which can trigger plant cell death and then inhibit the cloning and spreading of phytoplasma in plant cell. However, the accumulation of anthocyanins can dampen the protective effect of oxygen radical and reduce the quantity of H2O2 that accumulated. As a consequence, the defense mechanism of plant was repressed, which may lead to an accumulation of the phytoplasma. This finding is in line with our results that the concentration of anthocyanins increased in PIS compared with HS. Similar results also were reported in phytoplasma infected grapevine, Arabidopsis thaliana, and Lasiodiplodia theobromae-infected peach (Hren et al., 2009; Scarpari et al., 2005). These results indicated that anthocyanin accumulation may suppress the plant defense systems, possibly resulting in high phytoplasma titers and then the death of the diseased host.
3.3 Changes of hormones responding to phytoplasma infectionHormonal imbalances triggered by phytoplasma infection have been found in previous studies and were considered involved in the formation of symptoms such as witches' broom and short internodes (Mou et al., 2013; Fan et al., 2104; Gai et al., 2014; Scarpari et al., 2005). Increased cytokinin (zeatin) concentration has been demonstrated to be related to the formation of these symptoms. In our metabonomics analysis, it was found that phytohormone balance was disturbed in the interaction of paulownia-phytoplasma, including zeatin and auxin. Zeatins, including cis-zeatin and trans-zeatin, are synthesized through isopentenyl pyrophosphate and play crucial roles in regulating the proliferation and differentiation of plant cells. Altering the concentration of cytokinins was responsible for lateral bud formation, shoots production, short internodes and may help the pathogen to invade the stele region and vasculature (Orchard et al., 1994; Moreau et al., 2014). In our study, the content of isopentenyl adenine, tran-zeatin, cis-zeatin and zeatin riboside increased significantly in PIS compared with healthy seedlings, which is in accord with previous report that the expression levels of several genes involved in zeatin biosynthsis were differentially expressed in phytoplasma infected seedlings at the transcriptome level, including IPT, CYP735A (Mou et al., 2013; Fan et al., 2014). Isopentenyl transferase (IPT) is the first and key enzyme in zeatin biosynthesis. Up-regulation of IPT may elevate the concentration of cytokinins. In many IPT transgenic plants, witches' broom and virescence were the most obvious symptoms (Xu et al., 2009; Hu et al., 2013). At the same time, key genes like CRE1 and AHP in CK metabolism were also up-regulated in our previous study at transcriptome level. These results suggested that high level of zeatin may be associated with witches' broom. Similar findings have also been reported in mulberry, Jujube, and peanut (Gai et al., 2014; Sun et al., 2008; Liu et al., 2014). Auxins have been reported to play significant role in anti-phytoplasma activity (Weintraub et al., 2010). In our study, the concentration of auxin changed slightly, thereby, increasing the cytokinin/auxin ratio, which may lead to the formation of lateral buds. In a word, this study further demonstrated that the overall effect of the differentially expressed hormones described above would be closely related to PaWB from metabonomic level.
4 ConclusionIn the present study, we preformed a metabolic analysis on healthy and phytoplasma-infected Paulownia tomentosa seedlings using the non-target HPLC-MS methods. Our result showed that 1 612 metabolites were differentially expressed among the two samples, KEGG pathway analysis revealed the candidate metabolites and inherent pathways potentially involved in PaWB infection. Our study will provide a better understanding of the molecular mechanisms that regulated PaWB and shed some light on the biochemical processes behind phytoplasma pathogenicity. Thus, the results of this study will help us to better understanding the plant-phytoplasma interactions and the mechanisms of Paulownia responses to phytoplasma infection.
| [] | Allwood J W, Ellis D I, Goodacre R. 2008. Metabolomic technologies and their application to the study of plants and plant-host interactions. Physiol Plantarum, 132(2): 117–135. |
| [] | Ates S, Ni Y, Akgul M, et al. 2008. Characterization and evaluation of Paulownia elongota as a raw material for paper production. Afr J Biotechnol, 7(22): 4153–4158. |
| [] | Bayliss K L, Saqib M, Dell B, et al. 2005. First record of'Candidatus Phytoplasma australiense' in Paulownia trees. Australas Plant Path(1): 123–124. |
| [] | Berli F J, Moreno D, Piccoli P, et al. 2010. Abscisic acid is involved in the response of grape (Vitis vinifera L.) cv. Malbec leaf tissues to ultraviolet-B radiation by enhancing ultraviolet-absorbing compounds, antioxidant enzymes and membrane sterols. Plant Cell Environ, 33(1): 1–10. |
| [] | Cao X B, Fan G Q, Deng M J, et al. 2014a. Identification of genes related to Paulownia witches' broom by AFLP and MSAP. Int J Mol Sci, 15(8): 14669–14683. DOI:10.3390/ijms150814669 |
| [] | Cao X B, Fan G Q, Zhai X Q. 2012. Morphological changes of the witches' broom seedlings of Paulownia tomentosa treated with methyl methanesulphonate and SSR analysis. Acta Phytopathologica Sin, 42(2): 214–218. |
| [] | Cao X B, Fan G Q, Zhao Z L, et al. 2014b. Morphological changes of paulownia seedlings infected phytoplasmas reveal the genes associated with witches' broom through AFLP and MSAP. PLoS One, 9(10): e112533. |
| [] | DiLeo M V, Strahan G D, den Bakker M, et al. 2011. Weighted correlation network analysis (WGCNA) applied to the tomato fruit metabolome. PLoS One, 6(10): e26683. DOI:10.1371/journal.pone.0026683 |
| [] | Durrant W E, Rowland O, Piedras P, et al. 2000. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. The Plant Cell Online, 12(6): 963–977. DOI:10.1105/tpc.12.6.963 |
| [] | Fan G Q, Dong Y P, Deng M J, et al. 2014. Plant-pathogen interaction, circadian rhythm, and hormone-related gene expression provide indicators of phytoplasma infection in Paulownia fortunei. Int J Mol Sci, 15(12): 23141–23162. DOI:10.3390/ijms151223141 |
| [] | Fan G Q, Zhang S, Zhai X Q, et al. 2007. Effects of antibiotics on the Paulownia witches' broom phytoplasmas and pathogenic protein related to witches' broom symptom. Sci Silv Sin, 43(3): 138–142. |
| [] | Gai Y P, Han X J, Li Y Q, et al. 2014. Metabolomic analysis reveals the potential metabolites and pathogenesis involved in mulberry yellow dwarf disease. Plant Cell Environ, 37(6): 1474–1490. DOI:10.1111/pce.2014.37.issue-6 |
| [] | Gould K S, McKelvie J, Markham K R. 2002. Do anthocyanins function as antioxidants in leaves? Imaging of H2O2 in red and green leaves after mechanical injury. Plant Cell Environ, 25(10): 1261–1269. DOI:10.1046/j.1365-3040.2002.00905.x |
| [] | Hren M, Nikolic P, Rotter A, et al. 2009. 'Bois noir' phytoplasma induces significant reprogramming of the leaf transcriptome in the field grown grapevine. BMC Genomics, 10(1): 460. DOI:10.1186/1471-2164-10-460 |
| [] | Hu J X, Tian G Z, Lin C L, et al. 2013. Cloning, expression and characterization of tRNA-isopentenyltransferase genes (tRNA-ipt) from paulownia witches'-broom phytoplasma. Acta Microbiologica Sinica, 53(8): 832–841. |
| [] | Ipekci Z, Gozukirmizi N. 2003. Direct somatic embryogenesis and synthetic seed production from Paulownia elongata. Plant Cell Rep, 22(1): 16–24. DOI:10.1007/s00299-003-0650-5 |
| [] | Ji X, Gai Y P, Zheng C, et al. 2009. Comparative proteomic analysis provides new insights into mulberry dwarf responses in mulberry (Morus alba L.). Proteomics, 9(23): 5328–5339. DOI:10.1002/pmic.v9:23 |
| [] | Jin K X, Liang C J, Deng D L. 1981. A study of the insect vectors of witches' broom in Paulownia trees. Linye Keji Tongxun(12): 23–24. |
| [] | Krall L, Huege J, Catchpole G, et al. 2009. Assessment of sampling strategies for gas chromatography-mass spectrometry (GC-MS) based metabolomics of cyanobacteria. J Chromatogr B, 877(27): 2952–2960. DOI:10.1016/j.jchromb.2009.07.006 |
| [] | Lee I M, Hammond R E, Davis R E, et al. 1993. Universal amplification and analysis of pathogen 16S rDNA for classification and identification of mycoplasmalike organisms. Phytopathology, 83(8): 834–42. DOI:10.1094/Phyto-83-834 |
| [] | Liu L Y, Tseng H I, Lin C P, et al. 2014. High-throughput transcriptome analysis of the leafy flower transition of Catharanthus roseus induced by peanut witches'-broom phytoplasma infection. Plant Cell Physiol, 55(5): 942–957. DOI:10.1093/pcp/pcu029 |
| [] | Liu R N, Dong Y P, Fan G Q, et al. 2013. Discovery of genes related to witches broom disease in Paulownia tomentosa×Paulownia fortunei by a De Novo assembled transcriptome. PLoS One, 8(11): e80238. DOI:10.1371/journal.pone.0080238 |
| [] | Luan H, Chen X M, Zhong S L, et al. 2013. Serum metabolomics reveals lipid metabolism variation between coronary artery disease and congestive heart failure: a pilot study. Biomarkers, 18(4): 314–321. DOI:10.3109/1354750X.2013.781222 |
| [] | Margaria P, Ferrandino A, Caciagli P, et al. 2014. Metabolic and transcript analysis of the flavonoid pathway in diseased and recovered Nebbiolo and Barbera grapevines (Vitis vinifera L. ) following infection by Flavescence doree phytoplasma. Plant Cell Environ, 37(9): 2183–2200. |
| [] | Moreau S, Fromentin J, Vailleau F, et al. 2014. The symbiotic transcription factor MtEFD and cytokinins are positively acting in the Medicago truncatula and Ralstonia solanacearum pathogenic interaction. New Phytol, 201(4): 1343–1357. DOI:10.1111/nph.12636 |
| [] | Mou H Q, Lu J, Zhu S F, et al. 2013. Transcriptomic analysis of Paulownia infected by Paulownia witches'-broom phytoplasma. PLoS One, 8(10): e77217. DOI:10.1371/journal.pone.0077217 |
| [] | Namba S. 2002. Molecular biological studies on phytoplasmas. J Gen Plant Pathol, 68(3): 257–259. DOI:10.1007/PL00013086 |
| [] | Niu S Y, Fan G Q, Deng M J, et al. 2016. Discovery of microRNAs and transcript targets related to witches' broom disease in Paulownia fortunei by high-throughput sequencing and degradome approach. Mol Genet Genomics, 291(1): 181–191. DOI:10.1007/s00438-015-1102-y |
| [] | Orchard J, Collin H, Hardwick K, et al. 1994. Changes in morphology and measurement of cytokinin levels during the development of witches' brooms on cocoa. Plant Pathol, 43(1): 65–72. DOI:10.1111/PPA.1994.43.issue-1 |
| [] | Roessner U, Wagner C, Kopka J, et al. 2000. Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J, 23(1): 131–142. DOI:10.1046/j.1365-313x.2000.00774.x |
| [] | ( Rural Industry Business Services. 1998. Paulownia: a commercial overview. Queensland: Department of Primary Industries. |
| [] | Scarpari L M, Meinhardt L W, Mazzafera P, et al. 2005. Biochemical changes during the development of witches' broom: the most important disease of cocoa in Brazil caused by Crinipellis perniciosa. J exp bot, 56(413): 865–877. DOI:10.1093/jxb/eri079 |
| [] | Schneiderová K, Šmejkal K. 2015. Phytochemical profile of Paulownia tomentosa (Thunb). Steud Phytochem Rev, 14(5): 799–833. DOI:10.1007/s11101-014-9376-y |
| [] | Sun Q, Zhou G, Sun H, et al. 2008. Effect of auxins on in vitro propagation of sour jujube infected by witches' broom phytoplasma. International Jujube Symposium, 840: 303–308. |
| [] | Torres M A, Jones J D E, Dangl J F. 2006. Reactive oxygen species signalling in response to pathogens. Plant Physiol, 141(2): 373–378. DOI:10.1104/pp.106.079467 |
| [] | Ward J L, Baker J M, Beale M H. 2007. Recent applications of NMR spectroscopy in plant metabolomics. FEBS J, 274(5): 1126–1131. DOI:10.1111/j.1742-4658.2007.05675.x |
| [] | Wang J, Zhang X, Shi M, et al. 2014. Metabolomic analysis of the salt-sensitive mutants reveals changes in amino acid and fatty acid composition important to long-term salt stress in Synechocystis sp. PCC 6803. Funct Integr Genomic, 14(2): 431–440. DOI:10.1007/s10142-014-0370-7 |
| [] | Want E J, Wilson I D, Gika H, et al. 2010. Global metabolic profiling procedures for urine using UPLC-MS. Nature Protocols, 5(6): 1005–1018. DOI:10.1038/nprot.2010.50 |
| [] | ( Weintraub P G, Jones P. 2010. Phytoplasmas: genomes, plant hosts and vectors. Wallingford: CABI. |
| [] | Wilson I D, Plumb R, Granger J. 2005. HPLC-MS-based methods for the study of metabonomics. J Chromatogr B, 817(1): 67–76. DOI:10.1016/j.jchromb.2004.07.045 |
| [] | Xu S, Sun Y M, Jia Z C, et al. 2009. Effect of overexpression of IPT and KN1 on development and growth of transgenic tobacco plants. Plant Physiol Commun, 45(6): 537–543. |
| [] | Zhang Y Z, Cao X B, Zhai X Q, et al. 2009. Study on DNA extraction of AFLP reaction system for Paulownia plants. J Henan Agric Univ, 43(6): 610–614. |
| [] | Zipfel C, Robatzek S. 2010. Pathogen-associated molecular pattern-triggered immunity: veni, vidi...?. Plant Physiol, 154(2): 551–554. DOI:10.1104/pp.110.161547 |
 2017, Vol. 53
2017, Vol. 53