文章信息
- 游韩莉, 袁义杭, 李长江, 张凌云
- You Hanli, Yuan Yihang, Li Changjiang, Zhang Lingyun
- 青杄MYB转录因子基因PwMYB20的克隆及表达分析
- Cloning and Expression Analysis of MYB Homologous Gene PwMYB20 from Picea wilsonii
- 林业科学, 2017, 53(5): 23-32.
- Scientia Silvae Sinicae, 2017, 53(5): 23-32.
- DOI: 10.11707/j.1001-7488.20170504
-
文章历史
- 收稿日期:2016-12-29
- 修回日期:2017-02-21
-
作者相关文章
转录因子是指能特异性结合靶基因启动子上顺式作用元件的蛋白质,对转录起着激活或抑制作用。在植物生长发育及抗逆调控网络中,转录因子发挥了十分重要的作用(Broun, 2004; Wang et al., 2016)。MYB家族转录因子是一类含有长为50~53个氨基酸残基且高度保守的MYB结构域的转录因子,是植物转录因子中最大的转录因子家族。Paz-Ares等(1987) 在玉米(Zea mays)中鉴定出第1个植物MYB基因,并命名为C1。之后,越来越多的MYB相关基因被分离鉴定。
MYB转录因子在结构上具有由1~4个MYB重复单元(R)组成的MYB结构域。根据R的个数,可以将其分为4个亚类,即只含1个R的MYB-related类型、包含2个R的R2R3-MYB类型、包含3个R的3R-MYB类型以及包含4个R的4R-MYB类型(Stracke et al., 2001; Dubos et al., 2010)。在植物中,R2R3-MYB类型MYB蛋白数量较多(Chen et al., 2006),且参与了植物的初生和次生代谢、植物细胞形态和模式建成、植物生长发育等多个生命过程的调节(Dubos et al., 2010; Ambawat et al., 2013)。拟南芥(Arabidopsis thaliana)中MYB11、MYB12和MYB111通过调控FLAVONOL SYNTHASE1等下游靶基因,调节类黄酮的生物合成(Stracke et al., 2007)。在许多物种中均鉴定到参与细胞壁形成的MYB转录因子,例如小麦(Triticum aestivum)的TaMYB4和玉米的ZmMYB31(Fornale et al., 2010; Ma et al., 2011)。此外,MYB对种子胚的发育、根系的发育等均有调控作用(Tominaga et al., 2008; Yang et al., 2009)。
此外,研究发现MYB家族基因也参与了植物逆境响应过程。干旱胁迫能显著诱导拟南芥AtMYB2的表达,在植物受到干旱胁迫时,AtMYB2可以激活一些受ABA诱导表达基因的转录,从而提高对干旱胁迫的耐受程度(Urao et al., 1993; Abe et al., 2003)。AtMYB44是一个R2R3类型的MYB转录因子,参与调节植物气孔闭合和植物对非生物胁迫的响应(Jung et al., 2008)。进一步研究发现,AtMYB44通过直接与WRKY70结合,抑制茉莉酸介导的防卫反应,激活水杨酸介导的防卫反应(Shim et al., 2013)。此外,水稻(Oryza sativa)中OsMYB2和小麦中的TaMYB4在植物响应生物胁迫与非生物胁迫中均发挥了重要作用(Yang et al., 2012; Al-Attala et al., 2014)。
青杄(Picea wilsonii)是松科(Pinaceae)云杉属(Picea)的一种常绿高大乔木,高可达50 m,是我国特有的针叶树种。其适应能力强,喜阴且极耐寒,多分布于凉爽湿润地区。目前,青杄已被多个地区列为水源涵养林及用材林的主要造林和更新树种。早期对青杄的研究多集中于青杄的群落生态学(张大勇等,1989),随后有研究者对青杄愈伤组织的诱导(杨映根等,1994)及育苗栽培(许家春等,2004)也进行了相关研究。近年来,随着分子生物学和细胞生物学的快速发展,对于青杄基因的功能研究也有了一定进展。本文通过RACE-PCR的方法,从青杄cDNA文库中克隆得到1个R2R3-MYB转录因子全长cDNA序列;采用荧光定量PCR分析其在青杄各组织以及各种非生物胁迫下的表达情况,并进一步利用亚细胞定位及转录激活活性验证试验揭示其生物学特性,为进一步探究其在植物生长发育及逆境响应中的功能打下基础, 同时有利于挖掘与利用木本植物中的优质基因。
1 材料和方法 1.1 试验材料及处理方法青杄花粉及种子均采集于北京植物园。3年生青杄幼苗的根、茎、针叶与花粉用于组织特异表达试验,将青杄种子播种于体积1:1蛭石和草炭土的培养基质中,置于温度21 ℃、相对湿度60%~70%、日照时间16 h的条件下培养,每周定时浇1次水。8周的青杄幼苗用于逆境响应试验。
逆境响应试验的处理方法参照张通等(2014)和李长江等(2014)略有改动:将8周的青杄幼苗裸根置于室温(25 ℃),于吸水纸上放置0,3,6,12 h;用100 mmol·L-1 NaCl处理青杄幼苗0,3,6,12 h;将青杄幼苗于清水中4 ℃处理0,3,6,12 h。为了验证PwMYB20对逆境胁迫的响应是否通过ABA途径,进行了外施ABA试验,即用100 μmol·L-1 ABA分别处理青杄幼苗0,3,6,12 h。
1.2 青杄PwMYB20全长cDNA的获得青杄cDNA文库通过Gateway方法构建,由Invitrogen(上海)公司完成(张盾等,2012)。在前期构建的多年生青杄均一化cDNA文库的基础上,利用RACE-PCR的方法得到PwMYB20末端序列,再经过序列拼接得到cDNA全长。RACE-PCR所用引物为MYB20-RACE-F、MYB20-RACE-R(表 1)。设计引物MYB20-F、MYB20-R从青杄cDNA文库克隆得到PwMYB20的ORF,连接到pEASY-T1上,获得PwMYB20单克隆。
|
|
PwMYB20生物信息学分析方法参照李长江等(2014)。利用DNAMAN软件进行PwMYB20的cDNA序列编码区预测及蛋白翻译。运用ProtParam工具(http://biopython.org/wiki/ProtParam)分析蛋白的理化特性和氨基酸组成。通过BLAST工具在NCBI(https://www.ncbi.nlm.nih.gov/)上进行核酸序列与蛋白序列的同源性分析。利用ClustalX软件进行氨基酸多序列比对分析,并用MEGA5软件基于邻位相连法构建系统发育树。利用WoLF-PSORT(http: //www.genscript.com/tools/wolf-psort)进行蛋白亚细胞定位预测。通过Protscale(http: //web.expasy.org/protscale/)、SignalP4.1(http: //www.cbs.dtu.dk/services/SignalP/)、TMHMM(http://www.cbs.dtu.dk/services/TMHMM/)、FoldIndex(http://bip.weizmann.ac.il/fldbin/findex)对蛋白疏水性、信号肽区域、跨膜结构域以及蛋白无序化位点进行预测分析。
1.4 PwMYB20组织特异性表达及逆境响应分析利用艾德莱(北京)公司的植物RNA快速提取试剂盒提取各试验材料的RNA,并用天根(北京)公司反转录试剂盒合成第1条cDNA链,置于-20 ℃保存。根据基因序列的非保守区设计定量引物为qMYB20-F、qMYB20-R。选取青杄EF1-α基因作为内参基因(李长江等,2014),引物为EF1-α-F、EF1-α-R。将合成的cDNA第1条链均一化浓度之后在StepOnePlus Real Time RT-PCR仪器上进行荧光定量PCR,引物序列详见表 1。试验分别设置2次生物学重复,3次技术重复。利用2-△△Ct法分析数据,用SPASS与SigmaPlot分析作图。
1.5 PwMYB20瞬时表达载体构建与亚细胞定位通过引物MYB20-F-KpnⅠ、MYB20-R-BamHⅠ(表 1)扩增PwMYB20编码区序列,扩增产物经双酶切后定向连入pEZS-NL载体(图 1)。构建好的表达载体转化大肠杆菌(Escherichia coli)菌株DH5α。利用天根(北京)质粒大型大量提取试剂盒提取质粒,质粒浓度至少达到1 μg·μL-1。通过基因枪将质粒轰击入洋葱(Allium cepa)表皮细胞(Yu et al., 2011)。将转化的洋葱表皮置于MS培养基上暗培养1天,用共聚焦显微镜(OLYMPUS,FV10i)观察拍照。为了确定PwMYB20表达部位,用空pEZS-NL载体所表达的绿色荧光以及DAPI荧光作为参照。

|
图 1 pEZS-NL的载体 Fig.1 Map of pEZS-NL 35S表示启动子序列;Ala10表示10个丙氨酸序列;EGFP表示GFP序列;Ocs3′表示章鱼碱合成酶基因的终止子。 35S shows promoter sequence; Ala10 shows 10 alanine sequences; EGFP shows GFP sequence; Ocs3′ shows terminator. |
设计引物,并在上游和下游引物分别加入EcoRⅠ和BamHⅠ酶切位点,将PwMYB20全长(1-225 aa)、N端(1-127 aa)和C端(133-225 aa)分别连入pGBKT7载体,引物详见表 1。构建好的表达载体转化大肠杆菌菌株DH5α,并测序验证。利用天根(北京)公司质粒小提试剂盒提取重组质粒,存于-20 ℃备用。将经测序验证正确后的重组质粒转入酵母AH109(Saccharomyces cerevisiae)菌株,操作按照上海唯地生物技术有限公司说明书进行。以转入空pGBKT7载体为阴性对照,转入pGBKT7-ANAC092为阳性对照(He et al., 2005),均匀涂于SD-Trp单缺陷平板上。生长2~3天后用接种针挑取少量的酵母单克隆,混于500 mL无菌水中,吸取5 μL滴在SD-Trp和含有10 mmol·L-1 3-氨基-1, 2, 4-三氮唑(3-AT)的SD-Trp-His-Ade缺陷型酵母培养基上。培养3~4天后,若在SD-Trp-His-Ade缺陷型酵母培养基上长出菌落,则具有转录激活活性,反之则无。
2 结果与分析 2.1 基因克隆与序列分析用RACE-PCR的方法得到末端序列,与EST序列拼接,得到PwMYB20的cDNA全长(图 2)。利用DNAMAN软件分析发现,PwMYB20的cDNA全长为966 bp,在86 bp处出现起始密码子ATG,在741 bp处出现终止密码子TGA,编码区共675 bp,编码225个氨基酸,在949 bp处出现PolyA尾巴。

|
图 2 PwMYB20的核酸序列与蛋白氨基酸序列 Fig.2 Nucleotide sequence and deduced protein amino acids sequence of PwMYB20 起始密码子(ATG)与终止密码子(TGA)用下划线标记,氨基酸序列用单字母表示,黑体字母表示PolyA尾巴。 Potential translation initiation codon (ATG) and termination codon (TGA) are underlined, amino acid residues are indicated by single letter code and PolyA tail are indicated by bold letters. |
利用ProtParam工具计算蛋白分子式为C1104H1740N340O330S8,分子量为25.3 kDa,等电点为9.11。亮氨酸(Leu)含量最高(9.8%),其次为8.9%的丝氨酸(Ser)与8.0%的精氨酸(Arg)。预测在体外半衰期为30 h,不稳定指数为55.02,说明蛋白不稳定。Protscale工具疏水性分析发现,苯丙氨酸(Phe81) 分值最大,为1.633,谷氨酰胺(Gln20) 分值最小,为-2.647。疏水位点与亲水位点均匀分布,推测该蛋白为亲水蛋白(图 3A)。SignalP工具预测发现该蛋白没有信号肽结构域(图 3B)。利用FoldIndex工具对蛋白质固有无序化进行分析,结果表明该蛋白固有无序化序列较多,推测在生理环境下蛋白的动态活性较大(图 3C)。TMHMM工具预测发现,整条肽链都位于膜外,因此推测该蛋白没有跨膜结构域(图 3D)。此外,利用WOLF-PSORT对PwMYB20亚细胞定位进行预测,发现其可能定位在细胞核中。

|
图 3 PwMYB20的理化特性分析 Fig.3 Physicochemical properties of PwMYB20 A:Protscale工具预测蛋白疏水性; B:PwMYB20的信号肽预测(C-score表示剪切位置分值,S-score表示信号肽分值,Y-score表示综合剪切位置分值); C:PwMYB20蛋白的固有无序化分析; D:PwMYB20的跨膜结构域分析, 图中第1条横线(1.0~1.2) 表示综合结果。 A: Hydrophobic analysis of PwMYB20; B: Signal peptide analysis of PwMYB20(C-score means Cleavage site score, S-score means Signal peptide score, Y-score means Combined cleavage site score); C: Intrinsically disordered protein analysis of PwMYB20; D: Transmembrane analysis of PwMYB20, the first horizontal line(1.0-1.2) in the graph represents the comprehensive result. |
通过BLAST工具在NCBI上用翻译后的蛋白序列进行检索,结果表明其属于R2R3-MYB家族,且在N端发现SANT结构域。选取拟南芥、白云杉(Picea glauca)等物种同源基因进行检索,发现PwMYB20与其他MYBs相似性较高。利用ClustalX工具进行多序列比对,发现PwMYB20与其他物种的MYBs在N端有较高的相似性,且为R2R3结构域,在C端则拥有比较特异的转录激活域TAD。与拟南芥相比,C端的结构域是青杄、白云杉等物种特有的(图 4)。

|
图 4 PwMYB20及同源蛋白的多序列比对
Fig.4 Multiple sequence alignment of PwMYB20 and homological proteins
线段示意不同结构域(R2, R3, TAD),虚线框示意青杄等物种中特有的结构域。相似性:黑色=100%;粉色≥75%;蓝色≥50%。 青杄Picea wilsonii:PwMYB20;白云杉Picea glauca:PgMYB10(ABQ51226.1),PgMYB5 (ABQ51221.1),PgMYB17 (ACN12959.1),PgMYB13(ABQ51229.1);火炬松Pinus taeda:PtMYB14(ABD60279.1);拟南芥Arabidopsis thaliana:AtMYB6 (NP_192684.1),AtMYB7(NP_179263.1),AtMYB32(NP_195225.1),AtMYB4(NP_195574.1). The lines indicate different domains (R2, R3, TAD), the dashed box indicates a specific domain in Picea wilsonii et al. Conserved percent: Black=100%; Pink≥75%; Blue≥50%. |
利用MEGA5软件的邻位相连法进行系统发育树的构建,可以发现PwMYB20与PgMYB20聚为一簇,拟南芥的MYBs与木本植物的MYBs明显分为两大类(图 5),进而推测2类的激活域TAD拥有不同的分子功能。

|
图 5 PwMYB20的系统发育树分析 Fig.5 Phylogenetic tree analysis of PwMYB20 MEGA5用于构建系统发育树,计算方法为邻位相连法。每个分支上的数字表示1 000次重复搜索的置信度。 MEGA5 is applied to construct the tree by Neighbor-joining method. Numbers on branches indicate bootstrap estimates for 1 000 replicate analysis. |
利用RT-qPCR试验检测PwMYB20在青杄各组织中的表达模式,结果发现PwMYB20在青杄的各个组织中均有表达,在种子中的表达量最高,其次是在针叶中,在花粉中的表达相对较少(图 6)。说明PwMYB20基因属于组成型表达,可能对青杄种子、针叶、花粉、茎和根的发育均有影响。
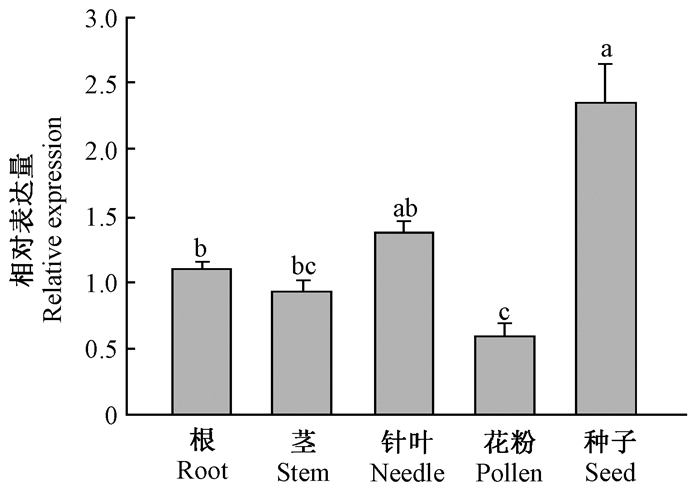
|
图 6 PwMYB20在青杄各组织的相对表达量 Fig.6 Expression analysis of PwMYB20 in different tissues of Picea wilsonii 利用单因素方差分析进行差异显著性分析,多重比较方法为Duncan(α=0.05) 法,不同字母表示差异显著(P < 0.05)。内参基因为青杄EF1-α。下同。 Single factor analysis of variance is used to analyze the difference, Duncan (α=0.05) test is used as the multiple comparison method, and different letters indicate significant difference (P < 0.05). EF1-α is the reference gene. The same below. |
为了研究不同胁迫条件下PwMYB20的表达模式,取胁迫处理后的整株青杄幼苗,提取RNA后进行反转录,浓度均一化后进行荧光定量PCR试验。结果表明,PwMYB20对干旱、4 ℃和ABA处理均有响应,而NaCl处理对PwMYB20表达的影响相对较弱(图 7)。在干旱处理下,PwMYB20表达量先上升后下降,在处理6 h后表达量最高,为未处理幼苗表达量的7倍。4 ℃低温处理3 h和12 h时PwMYB20的表达量显著高于未处理的幼苗,而4 ℃处理6 h时PwMYB20的表达量并没有明显上升,呈现上升—下降—上升的趋势。ABA处理显著提高了PwMYB20的表达量,其表达量持续上升,处理12 h后PwMYB20的表达量是未处理幼苗表达量的8倍。

|
图 7 不同处理下PwMYB20的相对表达量 Fig.7 Expression analysis of PwMYB20 with different treatments 内参基因为青杄EF1-α。各处理0 h时相对表达量为“1”。 EF1-α is the reference gene. The relative expression of PwMYB20 with different treatments for 0 h is '1'. |
pEZS-NL载体是CaMV 35S驱动的植物瞬时表达载体,具有GFP表达序列(图 1)。将PwMYB20构建在pEZS-NL载体上,与GFP融合表达,最终可以通过观察GFP的荧光确定PwMYB20的表达部位。结果显示,空载体GFP分布于整个细胞(图 8A, B, C),而PwMYB20与GFP的融合蛋白虽然在细胞质中也有微量表达(图 8D),但主要分布在细胞核中(图 8D, E, F)。这些结果表明PwMYB20是一个主要定位在细胞核中的蛋白。

|
图 8 PwMYB20的亚细胞定位(标尺:75 μm) Fig.8 Subcellular localization analysis of PwMYB20(Bar=75 μm) A, B, C:35S驱动GFP在洋葱(Allium cepa)中瞬时表达(A:空载体GFP荧光;B:明场下的洋葱表皮细胞;C:A与B的融合图像);D:PwMYB20-GFP在洋葱中瞬时表达;E:DAPI染色;F:GFP与DAPI荧光重叠。 A, B, C: Onion (Allium cepa) epidermal cells that are transformed with GFP driven by 35S promoter (A: The GFP fluorescence of empty plasm; B: Onion epidermal cell under bright field; C: The merged picture of A and B); D: Onion epidermal cell that is transformed with PwMYB20-GFP; E: Onion epidermal cell that is stained by DAPI; F: The overlap between GFP and DAPI fluorescence. |
为验证PwMYB20是否作为转录因子发挥作用,构建不同载体并转化酵母。图 9显示,转化空载体pBD和pBD-PwMYB20的酵母不能在-Trp-His-Ade缺陷型培养基中正常生长,而转化阳性对照pBD-ANAC092的酵母可以在-Trp-His-Ade缺陷型培养基中正常生长,表明PwMYB20全长没有转录激活活性。为了研究PwMYB20 N端和C端的激活活性,又分别构建了包含PwMYB20 N端和C端的酵母转化载体,结果显示:PwMYB20-133-225具有转录激活活性,而PwMYB20-1-127不具有转录活性(图 9)。这些结果表明包含TAD激活域的PwMYB20的C端具有激活活性,而全长没有转录激活活性,可能是由于N端存在转录抑制结构域。
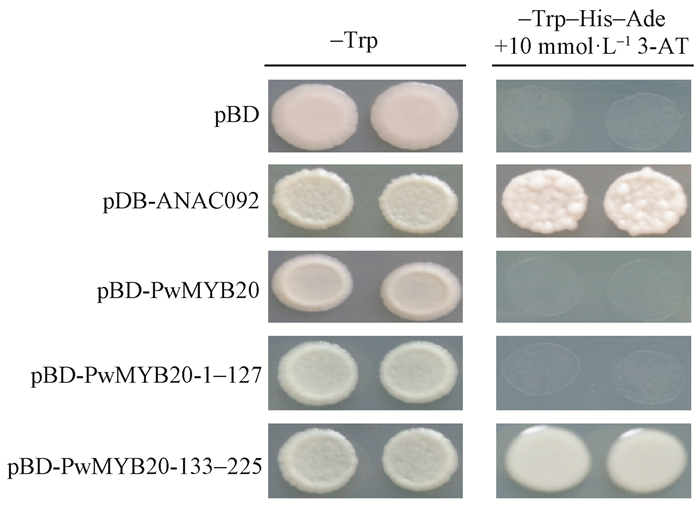
|
图 9 PwMYB20在酵母中的转录激活活性检测 Fig.9 Transcriptional activity test of the PwMYB20 proteins in yeast 空载体pBD和pBD-ANAC092分别作为阴性和阳性对照,图中蛋白后面的数字指示片段的位置。 The pBD vector alone and pBD-ANAC092 are used as negative and positive controls, and the numbers indicate the position of truncated fragments of protein. |
植物中的MYB基因最早在玉米中分离出来,被命名为C1(Paz-Ares et al., 1987),随后分离鉴定的MYB转录因子越来越多。拟南芥中已经发现198个MYB相关基因,而在水稻、玉米、二倍体粗山羊草(Aegilops tauschii)等植物中发现MYB基因均超过了200个。本试验从青杄中克隆得到PwMYB20基因,通过多序列对比发现,PwMYB20属于R2R3类型的MYB转录因子(图 4)。序列比对发现PwMYB20与其他物种的MYBs在N端有较高的相似性,且为R2R3结构域,在C端则拥有比较特异的转录激活域TAD。与拟南芥相比,C端的结构域是青杄、白云杉等物种特有的。R2R3-MYB类蛋白是植物中数目最多的一类MYB蛋白,拟南芥中198个MYB基因,其中编码R2R3-MYB的MYB基因数量高达126,占60%以上(Chen et al., 2006)。
本研究通过转录激活活性试验,发现PwMYB20全长及N端没有激活活性,而C端具有激活活性(图 9)。Hao等(2010)在研究NAC转录因子的激活活性时发现,NAC的N端存在1个NARD结构域,抑制了NAC的转录激活活性(Hao et al., 2010)。因此,推测PwMYB20 N端可能存在一个转录抑制区域,抑制了PwMYB20的转录激活活性。本文中亚细胞定位试验结果表明,PwMYB20主要存在于细胞核中(图 8),此结果与利用WoLF-PSORT工具进行亚细胞定位预测结果相符。因此推测PwMYB20作为一个转录因子主要在细胞核中发挥功能。
以往研究发现MYB转录因子在模式植物及作物类植物响应逆境胁迫中扮演了重要角色(Agarwal et al., 2006; Dai et al., 2007; Chen et al., 2013; Baldoni et al., 2015)。本文的研究结果显示,PwMYB20在青杄冷胁迫及干旱胁迫后表达量有显著变化(图 7),表明其可能在植物冷胁迫及干旱胁迫响应中发挥了功能。在低温处理3 h和12 h时PwMYB20表达量均显著高于未处理幼苗,而在低温处理6 h时PwMYB20表达量与未处理幼苗相比并没有显著变化,表明PwMYB20对于低温处理的响应存在时间上的差异,主要在处理早期和后期发挥作用。类似地,Shi等(2014)研究发现,AtHAP5A和AtXTH21在NaCl处理3,6,24 h时表达量均明显上升,而NaCl处理12 h时表达量却没有明显变化。
根据在植物抗逆胁迫过程中对ABA信号传导途径的依赖性,可将MYB转录因子分成2类:一类是和ABA信号相关的MYB转录因子,而另一类是独立于ABA信号途径的MYB转录因子。AtMYB60与AtMYB96可以通过ABA信号级联调节气孔运动(Cominelli et al., 2005),从而提高抗旱性和抗病性(Seo et al., 2009; 2010)。厚叶旋蒴苣苔(Boea crassifolia)中的BcMYB1对干旱胁迫响应显著,同时能被低温、PEG、高盐等胁迫诱导,但在外源ABA处理后其表达量却很低, 说明可能通过一种不依赖ABA的途径参与调控基因表达从而对逆境产生应答(Chen et al., 2005)。在本试验中,外源ABA处理时PwMYB20表达显著上升(图 7),因此推测PwMYB20可能通过ABA途径响应外界非生物胁迫,但其具体的调控与响应机制有待深入研究。
4 结论青杄PwMYB20,作为一个R2R3类型的MYB转录因子发挥作用,其转录激活活性位于C端。PwMYB20在青杄各组织中均有表达,属于组成型表达。此外,在干旱、低温和ABA处理下PwMYB20表达发生显著变化,说明其普遍参与了青杄应对逆境胁迫的响应过程。
| [] |
李长江, 崔晓燕, 孙帆, 等. 2014. 青杄干旱诱导基因PwWDS1的cDNA分离与表达分析. 林业科学, 50(4): 129–136.
(Li C J, Cui X Y, Sun F, et al. 2014. Isolation and expression analysis of PwWDS1 in Picea wilsonii. Scientia Silvae Sinicae, 50(4): 129–136. [in Chinese]) |
| [] |
许家春, 邵海燕, 李殿波. 2004. 优良绿化树种青杄云杉引种栽培技术. 中国林副特产(3): 24–25.
(Xu J C, Shao H Y, Li D B. 2004. Introduction and cultivation techniques of Picea wilsonii. Forest By-Product and Speciality in China(3): 24–25. [in Chinese]) |
| [] |
杨映根, 桂耀林, 唐巍, 等. 1994. 青杄愈伤组织在继代培养中的分化能力及染色体稳定性研究. 植物学报, 36(12): 934–939.
(Yang Y G, Gui Y L, Tang W, et al. 1994. Observation on differentiation potential and chromosome stability of callus in subculture of Picea wilsonii. Journal of Integrative Plant Biology, 36(12): 934–939. [in Chinese]) |
| [] |
张大勇, 赵松岭, 张鹏云, 等. 1989. 青杄林恢复演替过程中的邻体竞争效应及邻体干扰指数的改进模型. 生态学报, 9(1): 53–58.
(Zhang D Y, Zhao S L, Zhang P Y, et al. 1989. An improved model of neighborhood competition effect and neighborhood disturbance index in the process of restoration and succession in spruce forest. Acta Ecologica Sinica, 9(1): 53–58. [in Chinese]) |
| [] |
张盾, 刘亚静, 李长江, 等. 2012. 青杄均一化cDNA文库构建及EST序列分析. 生物技术通报(6): 71–76.
(Zhang D, Liu Y J, Li C J, et al. 2012. Construction of normalized cDNA library and analysis of corresponding EST sequences in Picea wilsonii. Biotechnology Bulletin(6): 71–76. [in Chinese]) |
| [] |
张通, 李巧玲, 张凌云. 2014. PwEXP1在青杄种子萌发及逆境响应中的表达特征. 林业科学, 50(12): 56–62.
(Zhang T, Li Q L, Zhang L Y. 2014. Expression characteristics of PwEXP1 gene in seed germination and adversity in Picea wilsonii. Scientia Silvae Sinicae, 50(12): 56–62. [in Chinese]) |
| [] | Abe H, Urao T, Ito T, et al. 2003. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. The Plant Cell, 15(1): 63–78. DOI:10.1105/tpc.006130 |
| [] | Agarwal M, Hao Y J, Kapoor A, et al. 2006. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. Journal of Biological Chemistry, 281(49): 37636–37645. DOI:10.1074/jbc.M605895200 |
| [] | Al-Attala M, Wang X, Abou-Attia M, et al. 2014. A novel TaMYB4 transcription factor involved in the defence response against Puccinia striiformis f. sp. tritici and abiotic stresses. Plant Molecular Biology, 84(4/5): 589–603. |
| [] | Ambawat S, Sharma P, Yadav N R, et al. 2013. MYB transcription factor genes as regulators for plant responses:an overview. Physiology and Molecular Biology of Plants, 19(3): 307–321. DOI:10.1007/s12298-013-0179-1 |
| [] | Baldoni E, Genga A, Cominelli E. 2015. Plant MYB transcription factors:Their role in drought response mechanisms. International Journal of Molecular Sciences, 16(7): 15811–15851. DOI:10.3390/ijms160715811 |
| [] | Broun P. 2004. Transcription factors as tools for metabolic engineering in plants. Current Opinion in Plant Biology, 7(2): 202–209. DOI:10.1016/j.pbi.2004.01.013 |
| [] | Chen B J, Wang Y, Hu Y L, et al. 2005. Cloning and characterization of a drought-inducible MYB gene from Boea crassifolia. Plant Science, 168(2): 493–500. DOI:10.1016/j.plantsci.2004.09.013 |
| [] | Chen Y, Chen Z L, Kang J Q, et al. 2013. AtMYB14 regulates cold tolerance in Arabidopsis. Plant Molecular Biology Reporter, 31(1): 87–97. DOI:10.1007/s11105-012-0481-z |
| [] | Chen Y H, Yang X Y, He K, et al. 2006. The MYB transcription factor superfamily of Arabidopsis:Expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology, 60(1): 107–124. DOI:10.1007/s11103-005-2910-y |
| [] | Cominelli E, Galbiati M, Vavasseur A, et al. 2005. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Current Biology, 15(13): 1196–1200. DOI:10.1016/j.cub.2005.05.048 |
| [] | Dai X Y, Xu Y Y, Ma Q B, et al. 2007. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiology, 143(4): 1739–1751. DOI:10.1104/pp.106.094532 |
| [] | Dubos C, Stracke R, Grotewold E, et al. 2010. MYB transcription factors in Arabidopsis. Trends in Plant Science, 15(10): 573–581. DOI:10.1016/j.tplants.2010.06.005 |
| [] | Fornale S, Shi X H, Chai C L, et al. 2010. ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant Journal, 64(4): 633–644. DOI:10.1111/tpj.2010.64.issue-4 |
| [] | Hao Y J, Song Q X, Chen H W, et al. 2010. Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta, 232(5): 1033–1043. DOI:10.1007/s00425-010-1238-2 |
| [] | He X J, Mu R L, Cao W H, et al. 2005. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant Journal, 44(6): 903–916. DOI:10.1111/tpj.2005.44.issue-6 |
| [] | Jung C, Seo J S, Han S W, et al. 2008. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiology, 146(2): 623–635. |
| [] | Ma Q H, Wang C, Zhu H H. 2011. TaMYB4 cloned from wheat regulates lignin biosynthesis through negatively controlling the transcripts of both cinnamyl alcohol dehydrogenase and cinnamoyl-CoA reductase genes. Biochimie, 93(7): 1179–1186. DOI:10.1016/j.biochi.2011.04.012 |
| [] | Paz-Ares J, Ghosal D, Wienand U, et al. 1987. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO, 6(12): 3553–3558. |
| [] | Seo P J, Park C M. 2010. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytologist, 186(2): 471–483. DOI:10.1111/j.1469-8137.2010.03183.x |
| [] | Seo P J, Xiang F N, Qiao M, et al. 2009. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiology, 151(1): 275–289. DOI:10.1104/pp.109.144220 |
| [] | Shi H T, Ye T T, Zhong B, et al. 2014. AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytologist, 203(2): 554–567. DOI:10.1111/nph.12812 |
| [] | Shim J S, Jung C, Lee S, et al. 2013. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant Journal, 73(3): 483–495. DOI:10.1111/tpj.12051 |
| [] | Stracke R, Ishihara H, Barsch G H A, et al. 2007. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant Journal, 50(4): 660–677. DOI:10.1111/j.1365-313X.2007.03078.x |
| [] | Stracke R, Werber M, Weisshaar B. 2001. The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology, 5(5): 447–456. |
| [] | Tominaga R, Iwata M, Sano R, et al. 2008. Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development, 135(7): 1335–1345. DOI:10.1242/dev.017947 |
| [] | Urao T, Yamaguchi-Shinozaki K, Urao S, et al. 1993. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. The Plant Cell, 5(11): 1529–1539. DOI:10.1105/tpc.5.11.1529 |
| [] | Wang H Y, Wang H L, Shao H B, et al. 2016. Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Frontiers in Plant Science, 7. DOI:10.3389/fpls.2016.00067 |
| [] | Yang A, Dai X Y, Zhang W H. 2012. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. Journal of Experimental Botany, 63(7): 2541–2556. DOI:10.1093/jxb/err431 |
| [] | Yang S W, Jang I C, Henriques R, et al. 2009. FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome a signals for inhibition of hypocotyl elongation. The Plant Cell, 21(5): 1341–1359. DOI:10.1105/tpc.109.067215 |
| [] | Yu Y L, Li Y Z, Huang G X, et al. 2011. PwHAP5, a CCAAT-binding transcription factor, interacts with PwFKBP12 and plays a role in pollen tube growth orientation in Picea wilsonii. Journal of Experimental Botany, 62(14): 4805–4817. DOI:10.1093/jxb/err120 |
 2017, Vol. 53
2017, Vol. 53


