文章信息
- 郭丽丽, 尹伟伦, 郭大龙, 侯小改
- Guo Lili, Yin Weilun, Guo Dalong, Hou Xiaogai
- 油用凤丹牡丹不同种植时间根际细菌群落多样性变化
- Variations of Bacterial Biodiversity in Rhizosphere Soils of Oil Tree Peony Cropping Continuously for Different Years
- 林业科学, 2017, 53(11): 131-141.
- Scientia Silvae Sinicae, 2017, 53(11): 131-141.
- DOI: 10.11707/j.1001-7488.20171115
-
文章历史
- 收稿日期:2017-08-07
- 修回日期:2017-10-17
-
作者相关文章
2. 北京林业大学生物科学与技术学院 北京 100083
2. College of Biosciences and Biotechnology, Beijing Forestry University Beijing 100083
油用牡丹(Paeonia Sect.Moutan)是结实能力强、可用来生产食用籽油的牡丹类型,产籽寿命长达30~50年(李育才,2015)。目前,在我国具有良好油用表现的主要是凤丹牡丹(Paeonia ostii)和紫斑牡丹(Paeonia rockii)。牡丹籽油富含亚麻酸等不饱和脂肪酸(Li et al., 2015a;2015b),同时含有白藜芦醇等药用成分(Chen et al., 2016; Mao et al., 2017),具有降血脂、改善神经功能、抑制癌细胞等功效(Su et al., 2016;Yu et al., 2017)。随着油用牡丹种植规模的扩大和种植时间的增加,牡丹植株呈现生长势逐年衰弱,开花量和产籽量降低,病虫害发生严重,甚至出现大片植株死亡等现象;同时,油用牡丹种苗基地培育牡丹种苗后,再次种植牡丹会出现种苗缓苗慢、生长势差、根系不发达等情况(马会萍等,2011)。连作障碍已成为限制油用牡丹产业发展的重要因素。
土壤微生物及其生态功能变化是土壤质量演变的关键因素(Beckers et al., 2017)之一,土壤根际微生物数量变化影响土壤养分的吸收和转化(Fierer et al., 2012; Zhang et al., 2014)。一种植物若种植年限过长,养分消耗过多,不利于养分平衡供给,造成土壤微生物细菌与真菌种群结构失衡,降低养分利用效率(Fierer et al., 2012;Qin et al., 2017;She et al., 2017;Tang et al., 2015;Zhang et al., 2014)。土壤微生物种群结构失衡是导致土壤质量下降,林木、作物、花卉等产生连作障碍的重要原因(Zhang et al., 2015;Fu et al., 2017;Tan et al., 2017;Chen et al., 2015)。根际微生物对促进作物生长、减少病原微生物侵害以及维持根际微生态平衡等具有重要作用(Li et al., 2014;Zhou et al., 2014;Dong et al., 2016; Xiong et al., 2015)。为了明确出现上述情况的原因,笔者对油用牡丹土壤根际细菌作了研究。
郑艳等(2016)采用Biolog ECO微平板和454焦磷酸测序技术研究了不同产区药用牡丹根际土栖真菌的活性变化,发现牡丹道地产区微生物整体活性高于非道地产区,根际土壤真菌在各产区呈现特异性分布,道地产区真菌系统发育相似性较高。韩继刚等(2016)发现部分拮抗牡丹病原菌的多黏类芽孢杆菌,该类牡丹根际微生物对其种子萌发和幼苗生长具有一定促生作用。Xue等(2014)和王雪山等(2012)采用DGGE技术分别鉴定了不同种植年限观赏牡丹根际土壤细菌群落结构变化。传统的土壤微生物研究方法有微生物平板培养法、Biolog鉴定系统法、生物标记法等,这些方法往往过低估计土壤微生物群落结构的组成,无法详细描述土壤微生物菌群的生理差异(赵帆等,2017)。
16S rRNA是原核生物核糖体30S小亚基的组成部分,其高变区基因序列随菌种亲缘关系不同有一定差异,是解释细菌物种间差异的特征核酸序列,可作为细菌系统发育和分类鉴定的指标,鉴定样本中的微生物种类(Pei et al., 2010)。以16S rRNA测序分析为平台的高通量测序技术,可一次性获得百万条16S rRNA序列,进行快速物种鉴定,具有样本量少、高通量和高精确性等优点(Beckers et al., 2017;Zarraonaindia et al., 2015;Rinke et al., 2013)。
目前关于油用牡丹土壤根际微生物菌群多样性变化的研究尚显薄弱,特别是随着种植年限的延长,根际微生物群落结构的动态变化特征不清。本研究以16S rRNA基因V3-V4区为分子标靶,采用高通量测序技术,分析不同种植年限油用凤丹牡丹根际细菌群落组成及多样性变化,以阐明其根际细菌群落结构随种植年限的变化规律,为解决牡丹连作障碍提供科学依据,为牡丹根际微生物资源的开发利用奠定基础。
1 材料与方法 1.1 研究区概况研究区设在洛阳国家牡丹园,位于洛阳市邙山中沟西(112°24′18.89″N,34.42′48.98″E),属亚热带季风型大陆气候,该区域海拔218~229 m,年均气温14.86 ℃左右,极端低温-15 ℃,极端高温42 ℃。年均降水量578.2 mm,年均蒸发量1 589.8 mm。区域内土壤以红黏土、黄褐土和褐土为主。调查发现,园区高年栽植的油用牡丹生长势及生产力出现一定程度的退化现象。
1.2 试验材料供试土壤取自洛阳国家牡丹园,园区坡度和耕作措施基本一致。选择种植年限分别为2、4、5、10和32年的油用凤丹牡丹根际土壤为研究对象,取样时间为种籽收获期(2016年8月)。每个种植年限按五点取样法随机选取3株长势一致的植株(每处理3个重复),每株以主茎为中心,半径约30~40 cm的范围取土,取样深度为0~20 cm,样品采用四分法混匀,去除土壤杂质后,分别装入无菌牛皮纸袋,放入冰盒带回实验室,-80 ℃保存。
1.3 试验方法 1.3.1 土壤微生物基因组DNA提取及质量检测采用FastDNA SPIN Kitfor Soil(USA)试剂盒提取不同种植年限的15个土壤样本的总DNA;采用1%琼脂糖电泳和Agilent 2100 Bioanalyzer检测DNA样品是否有降解以及杂质;NanoPhotometer(IMPLEN,德国)分光光度计检测DNA样品纯度;Qubit2.0 Flurometer检测DNA样品浓度;DNA样品于-20 ℃保存备用。
1.3.2 PCR扩增细菌16S rRNA基因V3-V4可变区取10 ng土壤基因组DNA为模板,使用TaKaRaEXtaq酶,以16S rRNA基因V3-V4可变区341F (5′-CCCTACACGACGCTCTTCCGATCTG(barcode) CCTACGGGNGGCWGCAG-3′)和805R (5′-GACTGG AGTTCCTTGGCACCCGAGAATTCCAGACTACHGGGT ATCTAATCC-3′)为引物,扩增16S rRNA基因序列V3-V4高变区,富集目的片段(Langille et al., 2013)。
1.3.3 16S rDNA文库质检及测序文库构建完成后,先使用Qubit 2.0进行初步定量;稀释文库至1 ng·μL-1,随后使用Agilent 2100对文库片段大小进行检测,片段大小符合预期则使用Bio-RAD CFX 96荧光定量PCR仪进行qPCR,对文库的有效浓度进行准确定量,以保证文库质量。检测合格的文库采用IlluminaMiSeq对16S rRNA基因序列的V3-V4区进行高通量测序。
1.3.4 信息分析流程去除测序所得序列中低质量碱基、接头污染序列,数据过滤后得到可信目标序列;根据末尾重叠情况将双端测序序列,利用算法PEAR进行序列拼接(Zhang et al., 2014);获得的标签与参考数据库中的OTUs序列进行比对,将相似度大于97%的序列归为一类OTUs,将OTUs比对到数据库中各物种相应的序列进行物种识别。利用QIIME 1.8.0软件对拼接后的序列进行OTUs交叠分析、聚类分析、系统发生树构建、Alpha多样性、Beta多样性等分析(Caporaso et al., 2010; Adler et al., 2013; Ahn et al., 2013)。
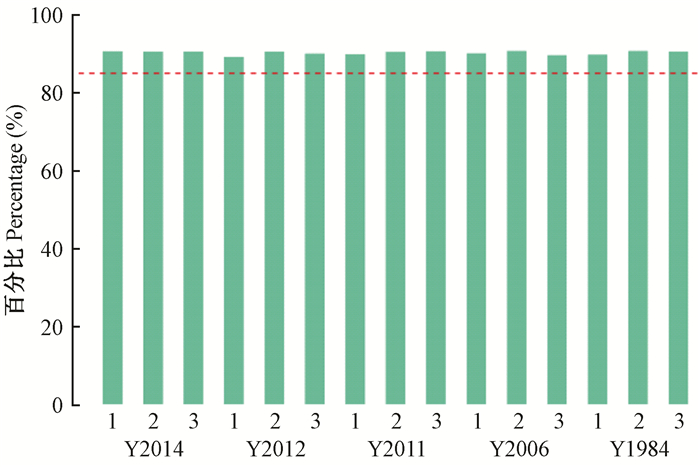
2 结果与分析 2.1 土壤样品测序深度评估油用凤丹牡丹根际细菌16S rRNA测序共获得432.1 Mb原始序列片段,过滤掉接头污染、低质量、含N比例大于5%的序列后,获得358.9 Mb Clean Reads(表 1)。为反映Clean Data质量,以Q30碱基百分比作为指标进行统计,Q30碱基百分比越大说明测序错误率小于0.1%的碱基在总碱基中的比例越大。图 1中所有样本Clean Reads的错误率均小于0.1%,Q30值均接近90%,说明测序质量或者建库质量较高。同时每个样本Q30碱基百分比非常相近,证明过滤后样本均一性较好。序列双端拼接共产生713 555条Tags,平均长度为449 bp。
|
|
|
图 1 Clean Reads质量统计 Figure 1 Distribution of Q30 rate 虚线表示Q30比例为85% Dot line represents that the Q30 was 85%. |
将油用凤丹牡丹15个根际土壤样本质控序列按97%相似性进行聚类,共获得2 366个OTUs,分属于24门,79纲、113目、117科、103属。所有土壤样本OUTs数目均介于800~950之间。2、4、5、10和32年生凤丹牡丹根际土壤样本分别获得2 729、2 628、2 656、2 650、2 595个OTUs,所获得的OUTs数目均一化较高(表 2)。
|
|
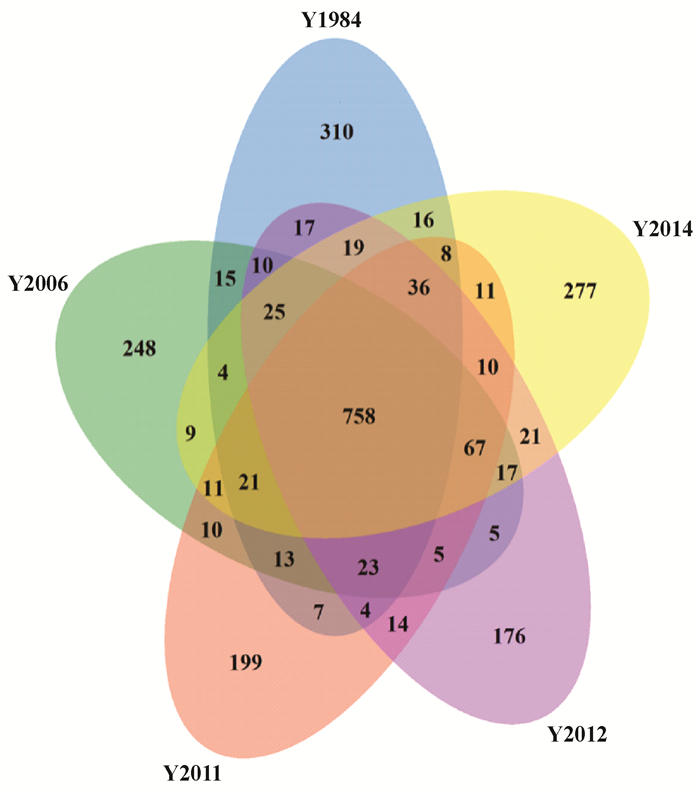
根据样品间OTUs交叠情况,分析不同种植年限共有和特有的OTUs数目,结合OTUs代表的物种,找出不同种植年限的核心及特异性微生物。结果表明,不同种植年限样品之间存在共有细菌OTUs数量为758个(图 2)。32年连作土壤样本特异性OTUs数量最多为310,2年为277,10年为248,5年和4年分别为199和176,表明不同种植年限凤丹牡丹根际细菌多样性存在一定差异。
|
图 2 不同种植年限油用凤丹牡丹土壤细菌群落OTUs交叠维恩图分析 Figure 2 Venn diagram of bacterial OTUs in rhizosphere soil resulted from the overlapping analysis of oil tree peony 'Fengdan' cropping continuously for different years |
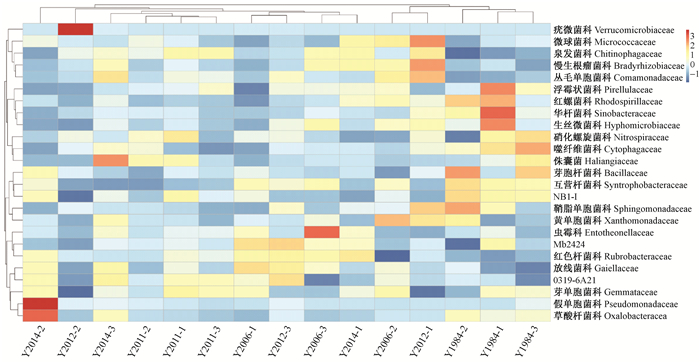
根据所有样品在科水平上的物种注释及丰度信息,选取丰度排名前25的科及其在每个样品中的丰度信息绘制热图,并从分类信息和样品间差异1个层面进行聚类,找出样品中聚集较多的物种(图 3)。结果表明,相同种植年限凤丹牡丹根际微生物样品基本上聚在一起,说明样品间重复性较好。其中32年生凤丹牡丹3个样品单独聚为一支,与其他年份间距离较远,说明它们之间的菌群丰度差异显著。从趋势上看,微生物菌群丰度随着种植年限的增加呈上升趋势,说明不同种植年限凤丹牡丹根际微生物菌群多样性存在差异。
|
图 3 不同种植年限油用凤丹牡丹根际土壤细菌物种丰度科水平上的聚类分析 Figure 3 Abundance and clustering analysis at family level for bacterial community in rhizosphere soil of oil tree peony 'Fengdan' cropping continuously for different years 横坐标为样品信息, 纵坐标为物种注释信息。图中左侧聚类树为物种聚类树;上方聚类树为样品聚类树;热图对应的值为每一行物种相对丰度经过标准化处理后得到的Z值。 X axes: Sample information. Y axes: OUTs Information. The left tree was from the clustering analysis of OUTs, and the up tree was from clustering analysis of samples. The corresponding value of the intermediate heat map is the Z-value obtained by the standardized treatment of the relative abundance of each row species. |
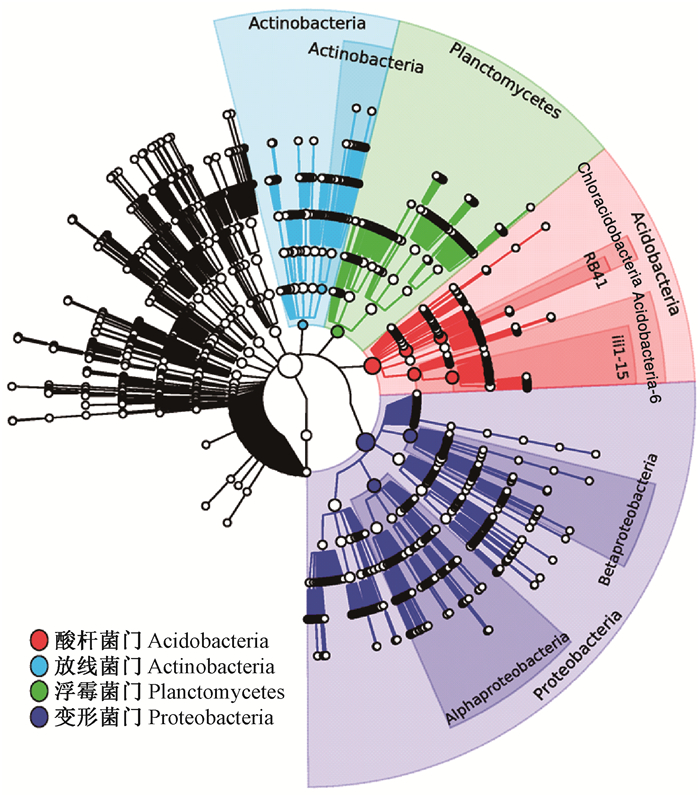
15个不同种植年限油用凤丹牡丹根际土壤样本的细菌群落微生物多样性的整体分布结果表明:根际土壤样本的中心优势细菌群落主要由放线菌门(Actinobacteria)、浮霉菌门(Planctomycetes)、酸杆菌门(Acidobacteria)、变形菌门(Proteobacteria)4个门类组成(图 4),说明以上4个门的细菌为凤丹牡丹根际土壤样本中较具优势的菌群。
|
图 4 油用凤丹牡丹根际土壤样本中的优势细菌群落组成分布情况 Figure 4 Distribution characteristics of predominant bacterial community composition in rhizosphere soil of oil tree peony 'Fengdan' cropping continuously for different years 不同颜色分别代表在整棵树里面比较重要的一些子树,包含序列越多圆圈越大。 Different colors represent more important subtrees in the whole tree. The more sequences it contains, the larger the circles will be. |
不同种植年限凤丹牡丹根际土壤样品中门水平下的中心优势菌种分析结果表明,变形菌门(26%)、酸杆菌门(11%)、浮霉菌门(10%)和放线菌门(7%)所占比例较高;拟杆菌门(Bacteroidetes)(4%)、厚壁菌门(Firmicutes)(3%)、疣微菌门(Verrucomicrobia)(3%)、芽单胞菌门(Gemmatimonadetes)(3%)、绿弯菌门(Chloroflexi)(2%)、硝化螺旋菌门(Nitrospirae)(1%)、装甲菌门(Armatimonadetes)(1%)、迷踪菌门(Elusimicrobia)(1%)、蓝藻门(Cyanobacteria)(1%)有一定占比(图 5)。

|
图 5 不同种植年限油用凤丹牡丹根际土壤细菌门水平上的群落结构特征 Figure 5 Bacterial community composition at phylum level in rhizosphere soil of oil tree peony 'Fengdan' cropping continuously for different years |
不同种植年限凤丹牡丹根际土壤中优势细菌群落门水平下的优势度指数结果表明:不同种植年限凤丹牡丹根际土壤中酸杆菌门、绿弯菌门、装甲菌门、迷踪菌门的优势度呈逐年上升趋势,其余细菌群落优势度呈逐年下降的趋势。不同种植年限凤丹牡丹根际土壤中酸杆菌门的优势度均高达41%以上。在32年生根际土壤样本中,酸杆菌门的优势度高达78.3%,装甲菌门的优势度较2年生增加近86%,并且检测到了特异性优势细菌门类(绿菌门和纤维杆菌门),且二者优势度均为0.1%(表 3)。32年生与小于10年生的凤丹牡丹根际土壤样本的比较分析发现,随着种植年限增加,除厚壁菌门的优势度变化不大外,其余根际优势细菌菌群多样性均呈下降趋势。其中,放线菌门、变形菌门、浮霉菌门、芽单胞菌门、硝化螺旋菌门等优势度下降趋势明显,放线菌门优势度下降比例高达90%,而疣微菌门、WS3、SBR1093及部分未知细菌甚至消失。推测随着种植年限的增加,酸杆菌门、绿弯菌门、装甲菌门等细菌的积累可能是导致凤丹牡丹根际细菌群落多样性下降的重要原因。
|
|
不同种植年限凤丹牡丹根际细菌群落在纲水平上的组成和优势菌属所占比例较为一致,隶属6个纲的细菌占据不同种植年限OTUs总数的30%以上。变形菌门的δ-变形菌纲(Deltaproteobacteria)(26%)、α-变形杆菌纲(Alphaproteobacteria)(25%)、β-变形菌纲(Betaproteobacteria)(15%)和γ-变形菌纲(Gammaproteobacteria)(15%);酸杆菌门的酸杆菌纲(Acidobacteria)(44%)和Solibacteres(14%);浮霉菌门的浮霉菌纲(Phycisphaerae)(27%)和Planctomycetia(60%);放线菌门的放线细菌纲(Actinobacteria)(25%)、酸微菌纲(Acidimicrobiia)(18%)、嗜热油菌纲(Thermoleophili)a(17%)、MB-A2-108(15%)、红杆菌纲(Rubrobacteria)(10%)为纲层次上的优势菌(图 6)。
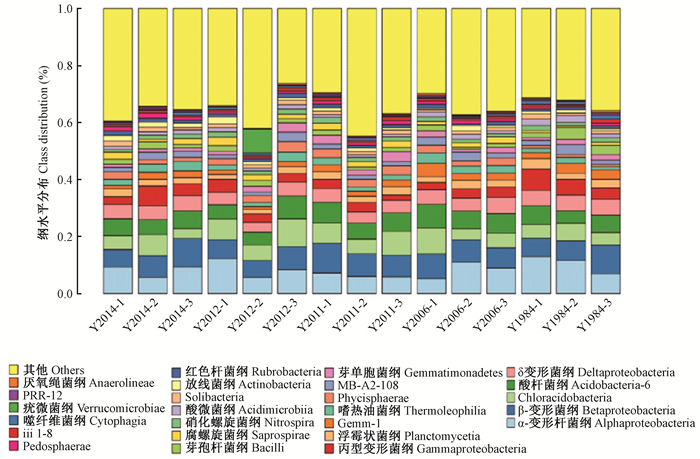
|
图 6 不同种植年限油用凤丹牡丹根际土壤细菌纲水平上的群落结构特征 Figure 6 Bacterial community composition at class level in rhizosphere soil of oil tree peony 'Fengdan' cropping continuously for different years |
Alpha多样性分析样品内菌种类别的丰富度和菌种数目的均匀度。Alpha多样性越高,细菌种类越丰富,群落越稳定。Shannon和Simpson指数评价群落物种组成的均匀度,Observed species和Chao1指数反映群落物种丰富度。不同种植年限间油用凤丹牡丹根际土壤样本中Shannon和Simpson指数表现出相同的变化规律,说明处理间差异不显著,不同样品均匀度较高。32年生凤丹牡丹根际土壤样本的Chao 1指数和Observed species指数与其他4个种植年限相比最小,说明连作降低了细菌群落多样性。因此,种植年限影响细菌的丰富度和多样性,但其影响程度在不同连作年限间差异较大(图 7)。
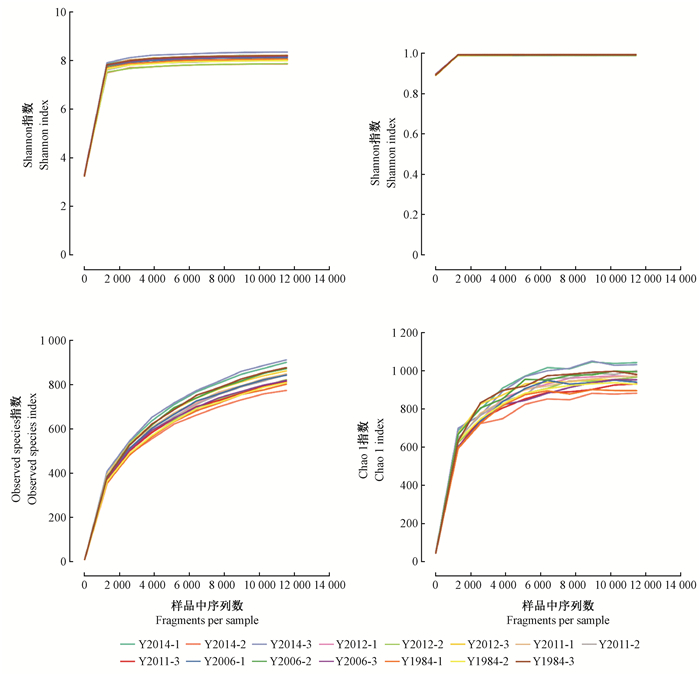
|
图 7 不同种植年限油用凤丹牡丹土壤微生物α多样性分析 Figure 7 α-diversity analysis of soil microbes in rhizosphere soil of oil tree peony 'Fengdan' cropping continuously for different years |
基于PCoA主坐标分析样品间菌群Beta多样性差异评估不同种植年限凤丹牡丹根际细菌群落的差异变化结果表明,不同种植年限土壤细菌群落在分布上存在差异,32年生土壤样本的细菌群落在PC1轴上的投影相对比较接近,全部分布在图 8的右边居中,且与图 8左方4个处理相距较远;左方4个年份细菌群落结构聚在一起,十分接近,说明种植年限影响凤丹牡丹根际土壤中细菌群落的变化(图 8)。
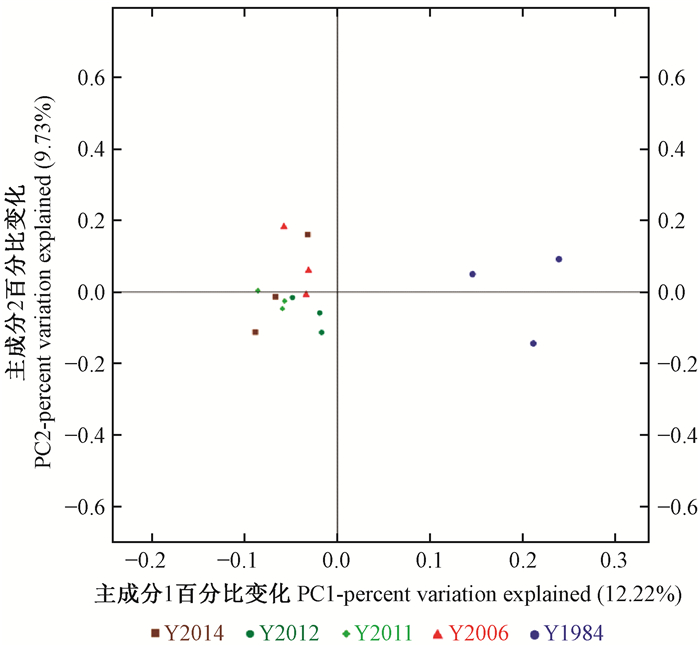
|
图 8 不同种植年限油用凤丹牡丹根际土壤细菌主坐标分析 Figure 8 Principal coordinate analysis of soil bacteria in rhizosphere soil of oil tree peony 'Fengdan' cropping continuously for different years |
连作障碍指同一种植物或近缘植物在同一地块上连续多年种植后,表现出生育状况变差、病虫害严重、产量降低、品质变劣的现象(Xiao et al., 2012)。连作障碍导致产量下降的原因主要是由于单一作物多年连续种植导致植物根际分泌物及植株残体随种植年限增加逐年积累,造成土壤微生物区系紊乱(Wang et al., 2015;Kessler et al., 2012;Qi et al., 2009)。根际微生态系统的失衡导致土传病原菌大量繁殖的同时抑制有益拮抗菌的生长,造成植物生长发育不良,影响产量及品质形成(Tan et al., 2017;董林林等,2017;纳小凡等,2016;Dong et al., 2016;Mazzola et al., 2012)。
根际微生物群落结构失调是牡丹连作障碍形成的重要因素(杨瑞先等,2017)。牡丹根系分泌物刺激或抑制微生物生长的次级代谢物质的产生,引起根际土壤理化性质的改变,导致根际微生物种群结构发生变化(史冬燕等,2013;覃逸明等,2009)。不同牡丹品种根际微生物的数量存在明显差异,观赏品种根际微生物数量少于药用品种(康业斌等,2006)。Xue等(2014)采用变性梯度凝胶电泳(DGGE)技术鉴定了5、12和25年生观赏牡丹根际土壤细菌群落结构变化,发现种植年限影响牡丹根际微生物群落结构变化,种植年限越长,其根际土壤中细菌和真菌群落结构多样性水平越低。王雪山等(2012)采用DGGE方法评价了3、5、8、12、20年的牡丹根际土壤微生物种群结构,发现牡丹根际土壤细菌多样性变化不显著,而真菌多样性水平随种植年限的增加而降低,菌群结构趋于简单。
本研究采用16S rRNA高通量测序技术,评价了2、4、5、10和32年生的油用凤丹牡丹根际细菌群落多样性变化,发现连续种植导致油用凤丹牡丹根际土壤细菌群落的中心优势菌种主要有变形菌门、酸杆菌门、浮霉菌门和放线菌门,土壤细菌优势菌群结构组成变化较小,但菌群多样性随种植年限的增加呈下降趋势。酸杆菌门等细菌群落的逐年积累,绿菌门和纤维杆菌门等特异性菌群的出现,放线菌门、变形菌门、浮霉菌门等菌群丰度的逐年降低,以及疣微菌门等菌群的消失,可能是造成油用凤丹牡丹连作障碍形成的重要原因。研究选用了种植年限长达32年的凤丹牡丹根际土壤样本,通过高通量测序的方法,对土壤微生物群落结构的组成及丰度差异进行了评价,对牡丹连作障碍形成原因的研究进行了补充。
研究表明酸杆菌门具有以叶绿素为基础的光合能力、降解植物残体多聚物、参与单碳化合物代谢等功能(Pankratov et al., 2011;Kanokratana et al., 2011)。接种放线菌菌剂,可促进丹参(Salvia miltiorrhiza)生长、提高丹参产量及抗病虫能力,调节丹参根域微生态平衡(段佳丽等,2015)。变形菌在硫氧化、碳固定及污水生物修复过程中发挥重要功能(Lenk et al., 2011;Padhi et al., 2013)。本研究发现不同种植年限油用凤丹牡丹根际土壤中的中心优势菌群中,酸杆菌门优势度呈逐年上升,放线菌门、变形菌门、浮霉菌门优势度逐年下降,推测随着种植年限的增加,微生物多样性的下降可能是导致油用凤丹牡丹连作障碍的主要原因之一。
4 结论油用凤丹牡丹根际土壤中酸杆菌门、绿弯菌门、装甲菌门、迷踪菌门等细菌群落的逐年积累,绿菌门和纤维杆菌门等特异性菌群的出现,放线菌门、变形菌门、浮霉菌门、芽单胞菌门、硝化螺旋菌门等菌群丰度的逐年降低及疣微菌门等菌群的消失,表明油用凤丹牡丹根际微生物对维持土壤根际微环境具有重要的生态学意义。油用凤丹牡丹根际真菌多样性及根际分泌物对根际微生物的促进或抑制作用尚需做进一步分析。本研究结果为解决牡丹连作障碍提供一定的科学依据,也为油用牡丹根际微生物资源的开发利用打下一定基础。
董林林, 牛玮浩, 王瑞, 等. 2017. 人参根际真菌群落多样性及组成的变化[J]. 中国中药杂志, 42(3): 443-449. (Dong L L, Niu W H, Wang R, et al. 2017. Changes of diversity and composition of fungal communities in rhizosphere of Panax ginseng[J]. China Journal of Chinese Materia Medica, 42(3): 443-449. [in Chinese]) |
段佳丽, 薛泉宏, 舒志明, 等. 2015. 放线菌Act12与腐植酸钾配施对丹参生长及其根域微生态的影响[J]. 生态学报, 35(6): 1807-1819. |
Duan J L, Xue Q H, Shu Z M, et al. Effects of combined application of actinomycetes Act12 bio-control agents and potassium humate on growth and microbial flora in rooting zone of Salvia miltiorrhiza Bge[J]. Acta Ecologica Sinica, 35(6): 1807-1819. |
韩继刚, 王云山, 胡永红. 2016. 拮抗多种牡丹病原菌的多粘类芽孢杆菌的应用[J]. 中国专利, CN105861378A. (Han J G, Wang Y S, Hu Y H. 2016. Application of bacillus subtilis strains against multiple peony pathogens[J]. China Patent, CN105861378A. [in Chinese]) |
康业斌, 商鸿生, 董苗菊. 2006. 凤丹与洛阳红根际微生物及其与根皮中丹皮酚含量的关系[J]. 西北农林科技大学学报:自然科学版, (12): 159-162. (Kang Y B, Shang H S, Dong M J. 2006. Relation between microbes in rhizosphere and paeonol content in root-bark in Fengdan and Luo yanghong[J]. Journal of Northwest A&F University:Natural Science Edition, (12): 159-162. [in Chinese]) |
李嘉珏, 张西方, 赵孝庆. 2011. 中国牡丹[M]. 北京: 中国大百科全书出版社. (Li J J, Zhang X F, Zhao X Q. 2011. Tree Peony of China[M]. Beijing: China Encyclopedia Press. [in Chinese]) |
李育才. 2015. 油用牡丹产业发展的思考[J]. 新产经, (6): 52-53. (Li Y C. 2015. Thinking about the development of oil in peony industry[J]. New Production, (6): 52-53. [in Chinese]) |
马会萍, 彭正锋. 2011. 牡丹重茬种植障碍产生的原因及对策[J]. 中国园艺文摘, 27(4): 122-123. (Ma H P, Peng Z F. 2011. The causes and countermeasures of planting obstacles for the planting of peony[J]. China Garden Digest, 27(4): 122-123. [in Chinese]) |
纳小凡, 郑国琦, 彭励, 等. 2016. 不同种植年限宁夏枸杞根际微生物多样性变化[J]. 土壤学报, 53(1): 241-252. (Na X F, Zheng G Q, Peng L, et al. 2016. The diversity of microbial diversity in the rhizosphere of Ningxia wolfberry was changed[J]. Acta Pedologica Sinica, 53(1): 241-252. DOI:10.11766/trxb201503030643 [in Chinese]) |
覃逸明, 聂刘旺. 2009. 药用牡丹的自毒作用及其防治措施[J]. 生物学杂志, 26(6): 76-79. (Qin Y M, Nie L W. 2009. The self-toxic effects of medicinal peony and its preventive measures[J]. Biology Journal, 26(6): 76-79. [in Chinese]) |
史冬燕, 王宜磊. 2013. 牡丹根际微生物区系及土壤酶活的研究[J]. 黑龙江农业科学, (6): 15-17. (Shi D Y, Wang Y L. 2013. Study on the flora and soil enzyme activity of the microflora of the peony root[J]. Heilongjiang Agricultural Sciences, (6): 15-17. [in Chinese]) |
王雪山, 杜秉海, 姚良同, 等. 2012. 种植年限对牡丹根际土壤微生物群落结构的影响[J]. 山东农业大学学报:自然科学版, 43(4): 508-516. (Wang X S, Du B H, Yao L T, et al. 2012. The effect of planting time on the microbiome structure of the mudan root soil[J]. Journal of Shandong Agricultural University:Natural Science Edition, 43(4): 508-516. [in Chinese]) |
杨瑞先, 王祖华, 赵梓轩, 等. 2017. 牡丹根际微生物的研究进展[J]. 河南农业科学, 46(6): 29-33. (Yang R X, Wang Z H, Zhao Z X, et al. 2017. The research progress of the microorganism of peony root[J]. Journal of Henan Agricultural Sciences, 46(6): 29-33. [in Chinese]) |
赵帆, 赵密珍, 王钰, 等. 2017. 不同连作年限草莓根际细菌和真菌多样性变化[J]. 微生物学通报, 44(6): 1377-1386. (Zhao F, Zhao M Z, Wang Y, et al. 2017. Biodiversity of bacteria and fungi in rhizosphere of strawberry with different continuous cropping years[J]. Microbiology China, 44(6): 1377-1386. [in Chinese]) |
郑艳, 刘炜, 黄军祥, 等. 2016. 基于牡丹根际土壤微生物的中药材道地性研究[J]. 药学学报, 51(8): 1325-1333. (Zheng Y, Liu W, Huang J X, et al. 2016. Study on the study of traditional Chinese medicinal materials based on the soil microorganism of peony root[J]. Acta Pharm Sin, 51(8): 1325-1333. [in Chinese]) |
Adler C J K, Dobney L S, Weyrich J, et al. 2013. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions[J]. Nature Genetics, 45(4): 450-455. DOI:10.1038/ng.2536 |
Ahn J R, Sinha Z, Pei C, et al. 2013. Human gut microbiome and risk for colorectal cancer[J]. Journal of the National Cancer Institute, 105(24): 1907-1911. DOI:10.1093/jnci/djt300 |
Beckers B M O, De Beeck N, Weyens W, et al. 2017. Structural variability and niche differentiation inthe rhizosphere and endosphere bacterial microbiome of field-grown poplar trees[J]. Microbiome, 5(1): 25. DOI:10.1186/s40168-017-0241-2 |
Caporaso J G J, Kuczynski J, Stombaugh K, et al. 2010. QⅡME allows analysis of high-throughput community sequencing data[J]. Nature Methods, 7(5): 335-336. DOI:10.1038/nmeth.f.303 |
Chen F X, Zhang Q, Zhang X, et al. 2016. Simultaneous synergistic microwave-ultrasonic extraction and hydrolysis for preparation of trans-resveratrol in tree peony seed oil-extracted residues using imidazolium-based ionic liquid[J]. Industrial Crops and Products, 94: 266-280. DOI:10.1016/j.indcrop.2016.08.048 |
Chen X S, Zhao J J, Yao Y Y, et al. 2015. Effects of bio-organic fertilizer and fungicide application on continuous cropping obstacles of cut chrysanthemum[J]. The Journal of Applied Ecology, 26(4): 1231-1236. |
Dong L J, Xu G, Feng X L, et al. 2016. Soil bacterial and fungal community dynamics in relation to Panaxnoto ginseng death rate in a continuous cropping system[J]. Scientific Reports, 6: 31802. DOI:10.1038/srep31802 |
Fierer N, Lauber C L, Ramirez K S, et al. 2012. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients[J]. Isme Journal, 6(5): 1007-1017. DOI:10.1038/ismej.2011.159 |
Fu H G, Zhang F, Zhang Z, et al. 2017. Effects of continuous tomato monoculture on soil microbial properties and enzyme activities in a solar greenhouse[J]. Sustainability, 9(2): 317. DOI:10.3390/su9020317 |
Kanokratana P, Uengwetwanit T, Rattanachomsri U, et al. 2011. Insights into the phylogeny and metabolic potential of a primary tropical peat swamp forest microbial community by metagenomic analysis[J]. Microbial Ecology, 61(3): 518-528. DOI:10.1007/s00248-010-9766-7 |
Kessler D S, Bhattacharya C, Diezel E, et al. 2012. Unpredictability of nectar nicotine promotes outcrossing by hummingbirds in Nicotiana attenuata[J]. The Plant Journal, 71(4): 529-538. DOI:10.1111/tpj.2012.71.issue-4 |
Langille M G I J, Zaneveld J G, Caporaso D, et al. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences[J]. Nature Biotechnology, 31(9): 814-821. DOI:10.1038/nbt.2676 |
Lenk S, Arnds J, Zerjatke K, et al. 2011. Novel groups of Gammaproteo bacteria catalyse sulfur oxidation and carbon fixation in a coastal, intertidal sediment[J]. Environmental Microbiology, 13(3): 758-774. DOI:10.1111/emi.2011.13.issue-3 |
Li J G, Ren G D, Jia Z J, et al. 2014. Composition and activity of rhizosphere microbial communities associated with healthy and diseased greenhouse tomatoes[J]. Plant and Soil, 380(1): 337-347. |
Li S L, Wang Q, Shu J, et al. 2015a. Fatty acid composition of developing tree peony (Paeonia Section Moutan DC.) seeds and transcriptome analysis during seed development[J]. BMC Genomics, 16(1): 208. DOI:10.1186/s12864-015-1429-0 |
Li S R, Yuan L, Chen L, et al. 2015b. Systematic qualitative and quantitative assessment of fatty acids in the seeds of 60 tree peony (Paeonia Section Moutan DC.) cultivars by GC-MS[J]. Food Chemistry, 173: 133-140. DOI:10.1016/j.foodchem.2014.10.017 |
Mao Y J, Han F, Tian X, et al. 2017. Chemical composition analysis, sensory, and feasibility study of tree peony seed[J]. Journal of Food Science, 82(2): 553-561. DOI:10.1111/jfds.2017.82.issue-2 |
Mazzola M, Manici L M. 2012. Apple replant disease:role of microbial ecology in cause and control[J]. Annual Review of Phytopathology, 50: 45-65. DOI:10.1146/annurev-phyto-081211-173005 |
Padhi S K, Tripathy S, Sen R, et al. 2013. Characterisation of heterotrophic nitrifying and aerobic denitrifying Klebsiella pneumoniae CF-S9 strain for bioremediation of wastewater[J]. International Biodeterioration & Biodegradation, 78: 67-73. |
Pankratov T A, Ivanova A O, Dedysh S N, et al. 2011. Bacterial populations and environmental factors controlling cellulose degradation in an acidic Sphagnum peat[J]. Environmental Microbiology, 13(7): 1800-1814. DOI:10.1111/j.1462-2920.2011.02491.x |
Pei A Y W E, Oberdorf C W, Nossa A, et al. 2010. Diversity of 16S rRNA genes within individual prokaryotic genomes[J]. Applied and Environmental Microbiology, 76(12): 3886-3897. DOI:10.1128/AEM.02953-09 |
Qi J J, Yao H Y, Ma X J, et al. 2009. Soil microbial community composition and diversity in the rhizosphere of a Chinese medicinal plant[J]. Communications in Soil Science and Plant Analysis, 40(9): 1462-1482. |
Qin S, Yeboah S, Xu X, et al. 2017. Analysis on fungal diversity in rhizosphere soil of continuous cropping potato subjected to different furrow-ridge mulching managements[J]. Frontiers in Microbiology: 8. DOI:10.3389/fmicb.2017.00845 |
Rinke C P, Schwientek A, Sczyrba N N, et al. 2013. Insights into the phylogeny and coding potential of microbial dark matter[J]. Nature, 499: 431-437. DOI:10.1038/nature12352 |
She S, Niu J, Zhang C, et al. 2017. Significant relationship between soil bacterial community structure and incidence of bacterial wilt disease under continuous cropping system[J]. Archives of Microbiology, 199(2): 267-275. DOI:10.1007/s00203-016-1301-x |
Su J, Ma C, Liu C, et al. 2016. Hypolipidemic activity of peony seed oil rich in α-linolenic, is mediated through inhibition of lipogenesis and upregulation of fatty acid β-oxidation[J]. Journal of Food Science, 81(4): H1001-H1009. DOI:10.1111/1750-3841.13252 |
Tan Y, Cui Y, Li H, et al. 2017. Rhizospheric soil and root endogenous fungal diversity and composition in response to continuous Panaxnoto ginseng cropping practices[J]. Microbiological Research, 194: 10-19. DOI:10.1016/j.micres.2016.09.009 |
Tang J, Xue Z, Daroch M, et al. 2015. Impact of continuous Salvia miltiorrhiza cropping on rhizosphere actinomycetes and fungi communities[J]. Annals of Microbiology, 65(3): 1267-1275. DOI:10.1007/s13213-014-0964-2 |
Wang M C, Wu Z C, Meng H, et al. 2015. Growth and physiological changes in continuously cropped eggplant (Solanum melongena L.) upon relay intercropping with garlic (Allium sativum L.)[J]. Frontiers in Plant Science: 6. DOI:10.3389/fpls.2015.00262 |
Xiao X Z, Cheng H, Meng M A, et al. 2012. Intercropping with garlic alleviated continuous cropping obstacle of cucumber in plastic tunnel[J]. Acta Agriculturae Scandinavica Section B Soil and Plant Science, 62(8): 696-705. |
Xiong W, Li Z, Liu H, et al. 2015. The effect of long-term continuous cropping of black pepper on soil bacterial communities as determined by 454 pyrosequencing[J]. Plos One, 10(8). DOI:10.1371/journal.pone.0136946 |
Xue D, Huang X. 2014. Changes in soil microbial community structure with planting years and cultivars of tree peony (Paeonia suffruticosa)[J]. World Journal of Microbiology & Biotechnology, 30(2): 389-397. |
Yu P, Wang L, Tang F, et al. 2017. Resveratrol pretreatment decreases ischemic injury and improves neurological function via sonic hedgehog signaling after stroke in rats[J]. Molecular Neurobiology, 54(1): 212-226. DOI:10.1007/s12035-015-9639-7 |
Zarraonaindia I S M, Owens P, Weisenhorn K W, et al. 2015. The soil microbiome influences grapevine-associated microbiota[J]. mBio, 6(2): e02527. |
Zhang H Z, Jiang L, Liu X, et al. 2015. Effects of intercropping on microbial community function and diversity in continuous watermelon cropping soil[J]. Fresenius Environmental Bulletin, 24(10A): 3288-3294. |
Zhang J K, Kobert T, Stamatakis A. 2014. PEAR:a fast and accurate Illumina Paired-End reAdmerge R[J]. Bioinformatics, 30(5): 614-620. DOI:10.1093/bioinformatics/btt593 |
Zhang X, Wei H, Chen Q, et al. 2014. The counteractive effects of nitrogen addition and watering on soil bacterial communities in a steppe ecosystem[J]. Soil Biology & Biochemistry, 72: 26-34. |
Zhou X, Gao D, Liu J, et al. 2014. Changes in rhizosphere soil microbial communities in a continuously monocropped cucumber (Cucumis sativus L.) system[J]. European Journal of Soil Biology, 60: 1-8. DOI:10.1016/j.ejsobi.2013.10.005 |
 2017, Vol. 53
2017, Vol. 53

