文章信息
- 宋志姣, 杨合宇, 翁启杰, 周长品, 李发根, 李梅, 卢万鸿, 罗建中, 甘四明
- Song Zhijiao, Yang Heyu, Weng Qijie, Zhou Changpin, Li Fagen, Li Mei, Lu Wanhong, Luo Jianzhong, Gan Siming
- 细叶桉群体的遗传多样性和受选择位点
- Genetic Diversity and Selective Loci in Eucalyptus tereticornis Populations
- 林业科学, 2016, 52(9): 39-47
- Scientia Silvae Sinicae, 2016, 52(9): 39-47.
- DOI: 10.11707/j.1001-7488.20160905
-
文章历史
- 收稿日期:2015-08-20
- 修回日期:2015-09-16
-
作者相关文章
2. 中国林业科学研究院热带林业研究所 热带林业研究国家林业局重点实验室 广州 510520;
3. 国家林业局桉树研究开发中心 湛江 524022;
4. 保山学院 保山 678000
2. Key Laboratory of State Forestry Administration on Tropical Forestry Research Research Institute of Tropical Forestry, Chinese Academy of Forestry Guangzhou 510520 ;
3. China Eucalypt Research Centre Zhanjiang 524022 ;
4. Baoshan University Baoshan 678000
桃金娘科(Myrtaceae)桉属(Eucalyptus)双蒴盖亚属(Symphyomyrtus)细叶桉(E. tereticornis)是优良的纸浆和人造板工业用材树种,在全球热带和亚热带地区广为引种(Eldridge et al.,1993)。细叶桉天然分布于巴布亚新几内亚南部和澳大利亚东海岸(9°—38°S)海边至内陆100 km的广大区域,海拔范围0~1 500 m,分布区包括多种气候类型、年降雨量500~1 500 mm(Eldridge et al.,1993)。相应地,细叶桉表型性状的变异较大,种源/家系试验表明其生长、干形、分枝和木材性质等性状均变异显著(王豁然等,1988;徐建民等,1993;2003;梁坤南等,1995;Chamshama et al.,1999;陆钊华等,2000;Varghese et al.,2009;Luo et al.,2014),一些种源甚至可耐受-7.9℃的冻害(林睦就等,2003)。但是,细叶桉分子水平上的变异却较少研究,只有Butcher等(2009)利用简单重复序列(simple sequence repeats,SSR)和限制性片断长度多态性(restriction fragment length polymorphism,RFLP)分析了3个群体,张党权等(2009)利用简单重复序列区间(inter-simple sequence repeats,ISSR)分析了4个小群体,更大范围乃至全分布区的群体多样性研究尚待开展。多样性水平的研究有助于阐明群体变异的范围和结构,对种质资源管理和育种策略制定亦有重要的参考价值(Yanchuk,2001)。
细叶桉广泛的分布区和原产地多种气候类型也为群体适应性研究提供了材料。适应性是生境条件对群体长期自然选择的结果,从而导致群体间基因频率的不同(Jump et al.,2006),进而引起表型和遗传结构的分化(Savolainen et al.,2007)。因此,阐明与自然选择有关的基因或基因组位点对理解适应性的遗传基础和指导种质资源的保护均有重要意义(Savolainen et al.,2013;Alfaro et al.,2014)。桉属中,目前仅在棒头桉(E. gomphocephala)(Bradbury et al.,2013)和毛果桉(E. tricarpa)(Steane et al.,2014)中研究了自然选择相关位点,2个树种分布范围均较窄(前者为西澳海岸30.4°—33.6°S,后者为南澳沿海36.7°—37.5°S),而对分布范围较广的树种却没有报道。本研究基于覆盖全基因组的44个基因组SSRs和64个开发自表达序列标签(expressed sequence tag,EST)的SSRs,利用25个中性的基因组SSRs分析9个细叶桉群体的遗传多样性和群体结构,利用全部标记分析受自然选择的候选位点,旨在明确该树种的遗传多样性和群体分化水平以及探索适应性的分子机制,并为种质资源管理和育种利用提供有用信息。
1 材料与方法 1.1 试验材料选取细叶桉9个天然群体的77个半同胞家系,每个家系选取1株生长正常的植株,各群体包含4~19株(表 1)。叶样采自2012年8月营建的、位于广东省遂溪县的细叶桉种源/家系试验林,参试家系的种子均采自天然林、母树间隔100 m以上,由澳大利亚林木种子中心和Kylisa种子公司提供。叶样采后临时保存在放有冰袋的泡沫盒中,带回实验室即置于-80℃冰箱备用。
|
|
称取0.3 g叶样放入1.5 mL Eppendorf管,液氮冷冻后通过球磨仪MM400(Retsch GmbH,德国)粉碎,采用改良CTAB法提取DNA(Gan et al.,2003)。利用NanoDrop 2000(Thermo Scientific,美国)测定DNA浓度和纯度。
基于已经发表的桉树SSR标记,选取在巨桉(E. grandis)全基因组上分布较均匀的108个SSR位点(包括44个基因组SSRs和64个EST-SSRs)(表 2)。考虑到所选SSR位点在基因组上相距较远,认为位点间不存在连锁不平衡。SSR引物由上海英骏生物技术有限公司合成。
|
|
SSR分型通过在10 μL PCR反应中加入荧光dUTP(Fermentas,加拿大)、采用降落PCR扩增程序、PCR产物利用ABI 3130xl测序仪(Applied Biosystems,美国)检测,具体参考Li等(2011),PCR反应的Mg2+浓度和扩增的退火温度视SSR标记而定。
1.3 数据分析 1.3.1 位点多样性分析对于各位点,利用Genepop 4.2软件(Rousset,2008)检测哈-温平衡(P ≤ 0.01,随机检测1 000次),利用PowerMarker 3.25(Liu et al.,2005)计算多态性信息量(polymorphism information content,PIC),利用GenAlEx 6.4.1(Peakall et al.,2006)计算等位片段数(number of alleles,Na)、观测杂合度(observed heterozygosity,Ho)、期望杂合度(expected heterozygosity,He)和固定指数(fixation index,F),利用FSTAT 2.9.3.2(http://www2.unil.ch/popgen/softwares/fstat.htm)计算等位片段丰富度(allelic richness,AR;基于最小群体的样品数)、群体间分化系数(between-population differentiation,FST)、群体内近交系数(inbreeding coefficients of individuals relative to the sub-population,FIS)和群体间近交系数(inbreeding coefficients of individuals relative to the total population,FIT),利用FreeNA(Chapuis et al.,2007)计算哑等位频率(null allele frequency,NAF)。
1.3.2 群体多样性分析因中性标记更适于群体分析(Black et al.,2001),只将既不偏离哈-温平衡、也在LOSITAN分析中(见下文)FST非异常值的25个中性的基因组SSR用于群体多样性和群体结构分析。
利用GenAlEx 6.4.1(Peakall et al.,2006)计算各群体Na,Ho,He和F等参数,利用FSTAT 2.9.3.2(http://www2.unil.ch/popgen/softwares/fstat.htm)计算AR(基于最小群体的样品数),手动统计特有等位片段数(number of private alleles,Npa)。
1.3.3 群体结构分析利用EXCEL计算25个中性位点对所有群体的FST,FIS和FIT平均值,利用GenAlEx 6.4.1(Peakall et al.,2006)进行分子方差分析(analysis of molecular variances,AMOVA);利用PowerMarker 3.25(Liu et al.,2005)计算群体间Nei’s遗传距离,采用非加权分组算术平均法(unweighted pair group method with arithmetic mean,UPGMA)进行聚类分析。
1.3.4 受选择位点的检测对所有108个SSRs,受选择位点的检测采用FST异常值和与气候因子关联分析2种方法。因部分群体较小,参考Prunier等(2011;2012)首先根据原产地的气候因子进行群体分组,从http://www.worldclim.org/(Hijmans et al.,2005)下载原产地1950—2000年的19个气候因子的数据,用R软件的cor.test功能检测气候因子之间的相关性,只取相关不显著的气候因子(P ≤ 0.05)进行9个群体的K-means分组(Legendre et al.,1998)。
根据各气候因子的群体分组,利用LOSITAN(Antao et al.,2008)通过FST与He的分布确定FST值异常的位点,选项设置为无限等位突变模型、模拟重复100 000次,FST值高于95%置信区间则为趋异选择(或正向选择)、低于5%置信区间则为平衡选择,假阳性率设为0.01
对于FST异常值的位点,再通过空间分析法(spatial analysis method,SAM)(Joost et al.,2007)检测等位频率与气候因子的关联,采用似然比值G和Wald检测(α=0.01)以及Bonferroni校正,关联显著性的置信水平为99.99%。显著关联位点的功能注释通过BlastX比对GenBank非冗余蛋白库(http://blast.ncbi.nlm.nih.gov/Blast.cgi;E ≤ 10-5)。为校正等位频率的自相关,通过等位片段在各群体的频率与气候因子的一元线性回归进一步验证关联的显著性(Narum et al.,2010)。
2 结果与分析 2.1 位点和群体多样性4 4个基因组SSR位点中,14个偏离哈-温平衡,6个显示FST值异常,其中1个同时偏离哈-温平衡和显示FST值异常(表 2),其余25个中性的基因组SSR用于群体多样性和群体结构分析。表 3列出了这25个位点对细叶桉9个群体的多样性参数,共检测到556个等位片段、平均每位点22.2个等位片段;位点多态性均较高,如PIC为0.680~0.954(平均0.904),按PIC ≥ 0.5的标准(Botstein et al.,1980)均为多态性高的位点。
|
|
表 4列出了细叶桉9个群体的SSR多样性参数。群体多样性水平都较高,如He为0.711~0.847(平均0.800)、AR为3.054~3.386(平均3.246)。各群体均有特有等位片段,数量为6~26(平均14.4)。但固定指数相对较高,平均达0.237,表明多数群体内存在一定程度的近交。
|
|
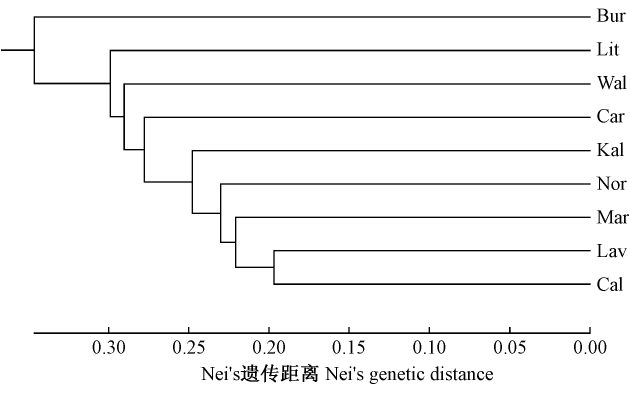
群体间分化水平较低,25个中性的基因组位点平均FST仅0.012(表 3);AMOVA分析也表明群体间方差分量仅占1.2%(表 5)。这表明细叶桉的群体分化水平较低。同时,25个位点的平均近交系数与AMOVA结果较一致,如FIS分别为0.316(表 3)与0.386(表 5)、FIT分别为0.324(表 3)和0.394(表 5),表明细叶桉群体变异主要存在于群体内。并且,聚类分析中9个群体逐步聚类(图 1),很难再区分亚类,也表明细叶桉具有较低分化的群体结构。
|
|
群体产地的19个气候因子中,年均气温、年均降水、最热月最高温度和季节性降水变异系数4个因子互不相关,用于K-means分组可将9个群体分别分为4,2,4和4组。FST异常值检测和SAM分析均基于气候分组进行。
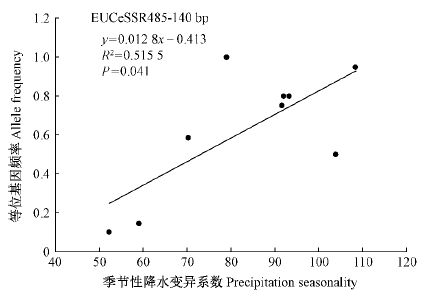
利用LOSITAN(Antao et al.,2008)共检测到78个(72.2%)FST值异常的位点(表 2),即为受选择的位点;年均气温、年均降水、最热月最高温度和季节性降水变异系数相关的FST值异常的位点数分别为27,10,51和42个(数据未显示),各气候因子上FST异常值大于95%置信水平的位点数分别为21,8,35和15个,而小于5%置信水平的位点数分别为6,2,16和27个。FST值异常的位点再进行SAM分析,发现其中4 个位点各有1个等位片段与1个或者2个气候因子显著关联(表 6)。功能注释表明:与季节性降水变异系数相关的EUCeSSR485位点与富含羟脯氨酸的蛋白家族[hydroxylproline-rich glycoprotein family protein(Vitis vinifera);2E-14]有关,与年均气温和年均降水均相关的位点EUCeSSR0497与跨膜内切1,4-β-葡聚糖酶[membrane-anchored endo-1,4-beta-glucanase(Gossypium hirsutum); 2E-12]基因同源,而EUCeSSR880和EUCeSSR1125没有明确的功能注释。进一步的线性回归分析也验证了其中1个等位片段(EUCeSSR485-140 bp)与季节性降水变异系数存在显著的线性关系(P ≤ 0.05)(图 2),而其余等位片段与相关气候因子的线性回归不显著,可能存在非线性关系。
|
|
|
图 2 EUCeSSR485-140 bp 频率与季节性降水变异系数的线性回归 Fig.2 Univariate linear regression of the frequency of EUCeSSR485-140 bp with precipitation seasonality |
细叶桉群体的遗传多样性水平高,基于25个中性的基因组SSR位点的He为0.711~0.847(平均0.800)(表 3)。这与Butcher等(2009)基于SSR分析3个细叶桉群体He为0.67~0.83的结果较一致,也与基于SSR分析的其他一些桉属树种的多样性水平类似,如尾叶桉(E. urophylla)群体的平均He为0.739(Payn et al.,2008)、棒头桉群体的平均He为0.75(Bradbury et al.,2013)、蓝桉(E. globulus)群体的平均He为0.82(Steane et al.,2006)。细叶桉较高的遗传多样性应是其分布范围广、异交系统和有效群体较大所致,这也是多数桉属树种的特点。
细叶桉的群体分化水平却偏低,平均FST仅0.012(表 3; 表 5)。这类似于尾叶桉FST为0.031(Payn et al.,2008),但显著低于同样分布广泛的赤桉(E. camaldulensis,FST=0.08)(Butcher et al.,2009)和蓝桉(FST=0.09)(Steane et al.,2006)。聚类分析(图 1)也表明细叶桉群体分化水平较低。这表明细叶桉较广的分布范围和不同的气候类型并未引起明显的遗传分化,其原因可能与连续分布、虫媒授粉和较为频繁的基因流有关。另一方面,尾叶桉(Payn et al.,2008)和本文的材料均是天然林采种育苗的子代、FST较低,而赤桉(Butcher et al.,2009)和蓝桉(Steane et al.,2006)的材料是天然林直接采集的植株样品,可能天然林的生境条件已在幼苗期对适应性差的迁移个体进行了选择、从而导致相对较高的群体分化参数(Kalisz et al.,2001)。
细叶桉群体的自然选择和适应可能涉及大量的基因组位点,一些位点具有明确的功能。基于不相关的4个气候因子的群体分组,共检测到FST值异常的78个SSR位点(72.2%),表明受选择的位点较多。虽然SAM分析只检测到其中4个位点与年均气温、年均降水和季节性降水变异系数的1~2个因子显著关联,但并不能认为其余
相对较小的群体仍可有效估算群体多样性参数。虽然本研究的多数群体较小、个体数不多,但与之前桉属树种中利用SSR估算的群体参数相近。类似地,Elliott等(2003)利用10个群体、每群体10株有效地进行了西方桉(E. occidentalis)的多样性分析。
本研究的结果对细叶桉种质资源管理和利用具有重要作用。资源保存和育种利用需要重视遗传多样性较高和特有等位片段数较多的群体,如Lav,Mar,Cal和Nor等群体,同时群体多样性水平普遍较高也表明育种利用的潜力较好。虽然细叶桉分子水平的群体遗传分化极低,但分子与表型的变异水平可能不同(Steane et al.,2006),之前的研究也表明细叶桉具有较大的表型变异(Varghese et al.,2009; Luo et al.,2014),因此育种中仍需进行种源/家系的表型选择,尤其是考虑到林木表型易受环境条件的影响。并且,因群体结构对标记与性状的关联分析具有较大影响,细叶桉低的群体分化水平表明该树种适于进一步进行标记与经济性状的关联研究。另外,检测到的受选择位点有助于理解细叶桉适应环境的分子机制,在当前气候变化背景下,这对探索多年生林木的环境适应潜力有积极作用。
4 结论本文首次系统地分析了细叶桉的群体多样性,检测了细叶桉与原产地气候因子显著关联的基因组位点,得到以下主要结论:1)细叶桉群体的遗传多样性高,育种利用的潜力大,种质资源管理应重视多样性较高和特有等位片段较多的群体; 2)细叶桉群体的遗传分化较低,其适于关联遗传分析; 3)鉴定了一批受选择位点,有助于理解林木适应环境的分子机制和探索林木环境适应性的潜力。
| [1] |
郭联华, 尹佟明, 李淑娴, 等. 2010. 植物细胞壁伸展蛋白研究述评. 林业科学 , 46 (12) : 144–152.
( Guo L H, Yin T M, Li S X, et al.2010. A review of extensins in plant cell wall. Scientia Silvae Sinicae , 46 (12) : 144–152. [in Chinese] ) (  0) 0)
|
| [2] |
梁坤南, 周文龙. 1995. 细叶桉种源-家系选择. 林业科学研究 , 8 (3) : 252–257.
( Liang K N, Zhou W L.1995. Study on selections of provenance and family for Eucalyptus tereticornis. Forest Research , 8 (3) : 252–257. [in Chinese] ) (  0) 0)
|
| [3] |
林睦就, 李柏海. 2003. 湖南桉树引种研究. 桉树科技 , 20 (1) : 1–12.
( Lin M J, Li B H.2003. The study on introduction of cold tolerant Eucalyptus in Hunan. Eucalypt Science and Technology , 20 (1) : 1–12. [in Chinese] ) (  0) 0)
|
| [4] |
陆钊华, 徐建民, 白嘉雨, 等. 2000. 细叶桉和赤桉种源间材性变异研究. 林业科学研究 , 13 (4) : 370–376.
( Lu Z H, Xu J M, Bai J Y, et al.2000. Study on wood property variation between Eucalyptus tereticornis and Eucalyptus camaldulensis. Forest Research , 13 (4) : 370–376. [in Chinese] ) (  0) 0)
|
| [5] |
王豁然, BanksJ C G, PryorL D. 1988. 细叶桉地理变异的多变量研究. 林业科学 , 24 (3) : 257–267.
( Wang H R, Banks J C G, Pryor L D.1988. A multivariate study of geographical variation in Eucalyptus tereticornis Sm. Scientia Silvae Sinicae , 24 (3) : 257–267. [in Chinese] ) (  0) 0)
|
| [6] |
徐建民, 陆钊华, 白嘉雨, 等. 2003. 细叶桉种源-家系综合选择的研究. 林业科学研究 , 16 (1) : 1–7.
( Xu J M, Lu Z H, Bai J Y, et al.2003. Study on integrated selection of provenances-families of Eucalyptus tereticornis. Forest Research , 16 (1) : 1–7. [in Chinese] ) (  0) 0)
|
| [7] |
徐建民, 吴坤明, 吴菊英, 等. 1993. 细叶桉地理种源生长性状遗传变异的分析与评价. 林业科学研究 , 6 (3) : 242–248.
( Xu J M, Wu K M, Wu J Y, et al.1993. An analysis on genetic variation in growth characters of geographical provenances of Eucalyptus tereticornis. Forest Research , 6 (3) : 242–248. [in Chinese] ) (  0) 0)
|
| [8] |
张党权, 田华, 谢耀坚, 等. 2009. 细叶桉遗传多样性的ISSR分析. 中南林业科技大学学报 , 29 (5) : 91–94.
( Zhang D Q, Tian H, Xie Y J, et al.2009. Genetic diversity of Eucalyptus tereticornis by ISSR. Journal of Central South University of Forestry&Technology , 29 (5) : 91–94. [in Chinese] ) (  0) 0)
|
| [9] |
周长品, 李发根, 翁启杰, 等. 2010. PCR产物直接测序和混合克隆测序进行桉树EST-SSR标记开发. 分子植物育种(网络版) , 8 (1) : e1.
( Zhou C P, Li F G, Weng Q J, et al.2010. Comparison between direct sequencing and pool-cloning-based sequencing of PCR products in EST-SSR marker development in Eucalyptus. Molecular Plant Breeding (online) , 8 (1) : e1. [in Chinese] ) (  0) 0)
|
| [10] |
Alfaro R I, Fady B, Vendramin G G, et al.2014. The role of forest genetic resources in responding to biotic and abiotic factors in the context of anthropogenic climate change. Forest Ecology and Management , 33 : 76–87.
( 0) 0)
|
| [11] |
Antao T, Lopes A, Lopes R J, et al.2008. LOSITAN:a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics , 9 : 323.
DOI:10.1186/1471-2105-9-323 ( 0) 0)
|
| [12] |
Black W C, Baer C F, Antolin M F, et al.2001. Population genomics:genome-wide sampling of insect populations. Annual Review of Entomology , 46 : 441–469.
DOI:10.1146/annurev.ento.46.1.441 ( 0) 0)
|
| [13] |
Botstein D, White R L, Skolnick M, et al.1980. Construction of genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics , 32 (3) : 314–331.
( 0) 0)
|
| [14] |
Bradbury D, Smithson A, Krauss S L.2013. Signatures of diversifying selection at EST-SSR loci and association with climate in natural Eucalyptus populations. Molecular Ecology , 22 (20) : 5112–5129.
DOI:10.1111/mec.12463 ( 0) 0)
|
| [15] |
Brondani R P V, Williams E R, Brondani C, et al.2006. A microsatellite-based consensus linkage map for species of Eucalyptus and a novel set of 230 microsatellite markers for the genus. BMC Plant Biology , 6 : 20.
DOI:10.1186/1471-2229-6-20 ( 0) 0)
|
| [16] |
Butcher P A, McDonald M W, Bell J C.2009. Congruence between environmental parameters,morphology and genetic structure in Australia's most widely distributed eucalypt,Eucalyptus camaldulensis. Tree Genetics&Genomes , 5 (1) : 189–210.
( 0) 0)
|
| [17] |
Chamshama S A O, Mugasha A G, Wate P A.1999. Variation in performance of Eucalyptus tereticornis provenances at Michafutene,Mozambique. Silvae Genetica , 48 (6) : 261–266.
( 0) 0)
|
| [18] |
Chapuis M P, Estoup A.2007. Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution , 24 (3) : 621–631.
( 0) 0)
|
| [19] |
Eldridge K, Davidson J, Harwood C, et al.1993. Eucalypt domestication and breeding. New York:Oxford University Press : 139–142.
( 0) 0)
|
| [20] |
Elliott C, Byrne M.2003. Genetic diversity within and between natural populations of Eucalyptus occidentalis (Myrtaceae). Silvae Genetica , 52 (3/4) : 169–173.
( 0) 0)
|
| [21] |
Gan S, Shi J, Li M, et al.2003. Moderate-density molecular maps of Eucalyptus urophylla S. T.Blake and E.tereticornis Smith genomes based on RAPD markers.Genetica , 118 (1) : 59–67.
( 0) 0)
|
| [22] |
He X, Wang Y, Li F, et al.2012. Development of 198 novel EST-derived microsatellites in Eucalyptus (Myrtaceae). American Journal of Botany , 99 (4) : e134–e148.
DOI:10.3732/ajb.1100442 ( 0) 0)
|
| [23] |
Hijmans R J, Camerson S E, Parra J L, et al.2005. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology , 25 (15) : 1965–1978.
DOI:10.1002/(ISSN)1097-0088 ( 0) 0)
|
| [24] |
Ito M, Kodama H, Komamine A, et al.1998. Expression of extensin genes is dependent on the stage of the cell cycle and cell proliferation in suspension-cultured Catharanthus roseus cells. Plant Molecular Biology , 36 (3) : 343–351.
DOI:10.1023/A:1005913818129 ( 0) 0)
|
| [25] |
Joost S, Bonin A, Bruford M W, et al.2007. A spatial analysis method (SAM) to detect candidate loci for selection:towards a landscape genomics approach to adaptation. Molecular Ecology , 16 (18) : 3955–3969.
DOI:10.1111/mec.2007.16.issue-18 ( 0) 0)
|
| [26] |
Jump A S, Hunt J M, Martinez-Izquierdo J A, et al.2006. Natural selection and climate change:temperature-linked spatial and temporal trends in gene frequency in Fagus sylvatica. Molecular Ecology , 15 (11) : 3468–3480.
( 0) 0)
|
| [27] |
Kalisz S, Nason J D, Hanzawa F M, et al.2001. Spatial population genetic structure in Trillium grandiflorum:the roles of dispersal,mating,history,and selection. Evolution , 55 (8) : 1560–1568.
DOI:10.1111/evo.2001.55.issue-8 ( 0) 0)
|
| [28] | Legendre P,Legendre L.1998.Numerical ecology.2nd ed.Amsterdam&New York:Elsevier. |
| [29] |
Li F, Gan S.2011. An optimised protocol for fluorescent-dUTP based SSR genotyping and its application to genetic mapping in Eucalyptus. Silvae Genetica , 60 (1) : 18–25.
( 0) 0)
|
| [30] |
Liu K, Muse S V.2005. PowerMarker:an integrated analysis environment for genetic marker analysis. Bioinformatics , 21 (9) : 2128–2129.
DOI:10.1093/bioinformatics/bti282 ( 0) 0)
|
| [31] |
Luo J, Arnold R, Lu W, et al.2014. Genetic variation in Eucalyptus camaldulensis and E. tereticornis for early growth and susceptibility to the gall wasp Leptocybe invasa in China.Euphytica , 196 (3) : 397–411.
( 0) 0)
|
| [32] |
Narum S R, Campbell N R, Kozfkay C C, et al.2010. Adaptation of redband trout in desert and montane environments. Molecular Ecology , 19 (21) : 4622–4637.
DOI:10.1111/mec.2010.19.issue-21 ( 0) 0)
|
| [33] |
Payn K G, Dvorak W S, Janse B J H, et al.2008. Microsatellite diversity and genetic structure of the commercially important tropical tree species Eucalyptus urophylla,endemic to seven islands in eastern Indonesia. Tree Genetics&Genomes , 4 (3) : 519–530.
( 0) 0)
|
| [34] |
Peakall R, Smouse P.2006. GenAlEx 6:genetic analysis in Excel. Population genetic software for teaching and research.Molecular Ecology Notes , 6 (1) : 288–295.
( 0) 0)
|
| [35] |
Prunier J, Gérardi S, Laroche J, et al.2012. Parallel and lineage-specific molecular adaptation to climate in boreal black spruce. Molecular Ecology , 21 (17) : 4270–4286.
DOI:10.1111/j.1365-294X.2012.05691.x ( 0) 0)
|
| [36] |
Prunier J, Laroche J, Beaulieu J, et al.2011. Scanning the genome for gene SNPs related to climate adaptation and estimating selection at the molecular level in boreal black spruce. Molecular Ecology , 20 (8) : 1702–1716.
DOI:10.1111/mec.2011.20.issue-8 ( 0) 0)
|
| [37] |
Rousset F.2008. Genepop'007:a complete reimplementation of the Genepop software for Windows and Linux. Molecular Ecology Resources , 8 (1) : 103–106.
DOI:10.1111/j.1471-8286.2007.01931.x ( 0) 0)
|
| [38] |
Savolainen O, Lascoux M, MeriläJ.2013. Ecological genomics of local adaptation. Nature Reviews Genetics , 14 (11) : 807–820.
DOI:10.1038/nrg3522 ( 0) 0)
|
| [39] |
Savolainen O, Pyhäjärvi T, Knürr T.2007. Gene flow and local adaptation in trees. Annual Review of Ecology,Evolution,and Systematics , 38 : 595–619.
DOI:10.1146/annurev.ecolsys.38.091206.095646 ( 0) 0)
|
| [40] |
Schlötterer C.2003. Hitchhiking mapping-functional genomics from the population genetics perspective. Trends in Genetics , 19 (1) : 32–38.
DOI:10.1016/S0168-9525(02)00012-4 ( 0) 0)
|
| [41] |
Shani Z, Dekel M, Tsabary G, et al.1997. Cloning and characterization of elongation specific endo-1,4-beta-glucanase (cel1) from Arabidopsis thaliana. Plant Molecular Biology , 34 (6) : 837–842.
DOI:10.1023/A:1005849627301 ( 0) 0)
|
| [42] |
Showalter A M.1993. Structure and function of plant cell wall proteins. Plant Cell , 5 (1) : 9–23.
DOI:10.1105/tpc.5.1.9 ( 0) 0)
|
| [43] |
Steane D A, Conod N, Jones R C, et al.2006. A comparative analysis of population structure of a forest tree,Eucalyptus globulus (Myrtaceae),using microsatellite markers and quantitative traits. Tree Genetics&Genomes , 2 (1) : 30–38.
( 0) 0)
|
| [44] |
Steane D A, Potts B M, Mclean E, et al.2014. Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Molecular Ecology , 23 (10) : 2500–1513.
DOI:10.1111/mec.12751 ( 0) 0)
|
| [45] |
Varghese M, Kamalakannan R, Harwood C E, et al.2009. Changes in growth performance and fecundity of Eucalyptus camaldulensis and E. tereticornis during domestication in southern India.Tree Genetics&Genomes , 5 (4) : 629–640.
( 0) 0)
|
| [46] |
Yanchuk A D.2001. A quantitative framework for breeding and conservation of forest tree genetic resources in British Columbia. Canadian Journal of Forest Research , 31 (4) : 566–576.
DOI:10.1139/x00-133 ( 0) 0)
|
| [47] |
Zhou C, He X, Li F, et al.2014. Development of 240 novel EST-SSRs in Eucalyptus L'Hérit. Molecular Breeding , 33 (1) : 221–225.
DOI:10.1007/s11032-013-9923-z ( 0) 0)
|
 2016, Vol. 52
2016, Vol. 52