文章信息
- 成龙平, 胡海涛, 郭卫东, 杨莉, 王长春, 杨玲
- Cheng Longping, Hu Haitao, Guo Weidong, Yang Li, Wang Changchun, Yang Ling
- 牛奶子实时定量PCR分析中内参基因的评价与验证
- Evaluation and Validation of Potential Reference Genes for Quantitative Real-Time PCR Analysis in Elaeagnus umbellata
- 林业科学, 2015, 51(5): 135-144
- Scientia Silvae Sinicae, 2015, 51(5): 135-144.
- DOI: 10.11707/j.1001-7488.20150516
-
文章历史
- Received date: 2014-07-21
- Revised date: 2014-10-03
-
作者相关文章
Elaeagnus umbellata is a rapid-growing large shrub that has been widely planted to prevent erosion and provide screening along highways due to its ability to fix nitrogen, drought and disease resistance, and tolerance of poor soil(Fordham et al., 2003). Most of the extracts from the flowers, leaves and berries display broad-spectrum antibacterial activity(Sabir et al., 2007). In addition, its red fruit is rich in minerals, vitamins, essential fatty acids, flavonoids and carotenoids, especially lycopene, which is about 5-20 times higher than that of ordinary tomato(Fordham et al., 2003; Ahmad et al., 2006; Guo et al., 2009). The sweet-tart fruit can be eaten fresh or processed for preserves, condiments, fruit-rolls and juice, or used as a substitute for tomato products(Fordham et al., 2003; Ahmad et al., 2006). Because of these unique features, E. umbellata has attracted considerable scientific interest, not only among nutritionist and agricultural scientists, but also ecologist and molecular biologists(Fordham et al., 2003; Ahmad et al., 2006; Guo et al., 2009). We have analyzed the expression changes of seven genes encoding enzymes of the main steps of carotenoid biosynthetic pathway during fruit ripening. The results revealed that the accumulation of lycopene in the fruit was concomitant with the up-regulation of upstream gene phytoene synthase(EutPsy) and the silence of downstream gene lycopene ε-cyclase(Guo et al., 2009). Therefore, the underst and ing of expression patterns of some key genes will help elucidate the mechanism involved in the specific physiological and biochemical process.
Quantitative real-time polymerase chain reaction(qRT-PCR)has rapidly become one of the most important tools in measuring gene expression levels due to its sensitivity, accuracy and specificity(Chao et al., 2012). However, to avoid experimental deviations or errors that inevitably occur during sample preparation and data analysis, all of which make quantitation of gene transcripts unreliable, normalization of the qRT-PCR data is essential. Among several normalization strategies, the use of one or more internal reference genes is currently the preferred way(Hong et al., 2008). An ideal reference gene should be stably expressed among various samples including those from developmental stages, organ types and external stimuli(Han et al., 2012). Housekeeping genes are routinely employed for this purpose, but these genes are not always stably expressed when tested in other species or under various experimental conditions. Recent studies have shown that no single reference gene is universal for all experiments, and using different reference genes can result in appreciable errors even up to 20-fold(Lu et al., 2013). Thus, it is crucial to systematically evaluate the expression stability of potential reference genes in a given set of biological samples from a specific organism.
To date, analysis of reference gene expression in plants has mainly focused on model and important crop species. E. umbellata belongs to the family Elaeagnaceae that is distant from any model plant species. Only a traditional housekeeping gene glyceraldehyde-3-phosphate dehydrogenase(GAPDH)has been used as reference gene(Guo et al., 2009; Su et al., 2013), but its stability has not yet been assessed in this species. Therefore, the objective of this study was to determine in systematic manner reference genes which demonstrate a high degree of expression stability under various organs, fruit ripening stages, hormone treatments and abiotic stress conditions in E. umbellata, to facilitate a more accurate normalization of qRT-PCR assays. The expression profiles of 12 potential reference genes including 14-3-3(Barsalobres-Cavallari et al., 2009), elongation factor 1-α(EF1-α)(Nicot et al., 2005; Hong et al., 2008; Silveira et al., 2009; Han et al., 2012; Zhu et al., 2013), eukaryotic initiation factor 4A(eIF4A)(Silveira et al., 2009; Lee et al., 2010; Zhu et al., 2012), RNA polymerase-Ⅱ(RPⅡ)(Tong et al., 2009), 60S ribosomal protein L7(RPL7)(Barsalobres-Cavallari et al., 2009), translation elongation factor2(TEF2)(Tong et al., 2009), ubiquitin-conjugating enzyme E2(UBCE)(Czechowski et al., 2005; Silveira et al., 2009), ubiquitin(UBQ)(Han et al., 2012; Fan et al., 2013), ubiquitin extension protein5(UBQ5)(Huis et al., 2010), 18S ribosomal RNA gene(18SrRNA)(Nicot et al., 2005), GAPDH(Hong et al., 2008; Barsalobres-Cavallari et al., 2009; Tong et al., 2009; Huis et al., 2010; Han et al., 2012) and β-actin(Actin)(Hong et al., 2008; Han et al., 2012)which were previously shown to have highly stable expression levels in other plant species, were evaluated in 29 samples. Five distinct statistical algorithms have been implemented to identify the best reference genes for qRT-PCR data normalization under different experimental conditions. Furthermore, the expression pattern of target gene EutPsy during fruit ripening was assessed using the selected reference genes.
1 Materials and methods 1.1 Plant materials and treatmentsFresh roots, stems, leaves, flowers and red fruits were harvested from 8-year-old trees grown under natural conditions at the experimental farm at Zhejiang Normal University, Jinhua, China. The fruits were sampled at green(approximately 19 weeks after anthesis), yellow(approximately 22 weeks after anthesis), dark yellow(approximately 24 weeks after anthesis), and full-ripe(approximately 27 weeks after anthesis)fruit ripening stages(Guo et al., 2009). For ABA and GA3 treatments, green fruits were dipping in 100 μmol ·L-1 solutions contained 0.2% Tween 80, respectively, and dried and sampled at 0, 6, 12 and 24 hours after hormone treatments(Tong et al., 2009). For heat treatment, the detached leaves with the developing leaf blade from 8-year-old trees were incubated at 40 ℃ for 8 h. The leaves were gathered at 2, 4, 6 and 8 h, respectively. For salt stress treatment, 4-week-old seedlings about 12 cm high in pots were supplemented with 300 mmol ·L-1 NaCl. The leaves, roots and stems were harvested at 0, 6 and 12 h, respectively. All 29 samples were collected in three replicates, immediately frozen in liquid nitrogen and stored at -80 ℃.
1.2 RNA extraction, quality control and cDNA synthesisTotal RNA was isolated according to a modified hot borate method(Guo et al., 2009) and digested with RNase-free DNaseⅠ(Takara, Toyoto, Japan). The purity of all RNA samples was assessed with a NanoDrop2000 spectrophotometer(Thermo, Wilmington, USA). Only RNA samples with an A260/ A280 ratio of 1.8-2.1 and an A260/A230 ratio of 2.0-2.3 were used for subsequent analysis. The RNA integrity was immediately checked using 1.5% agarose gel electrophoresis. The first-str and cDNA template was synthesized from2μg total RNA by M-MLV reverse transcriptase(Progema, Madison, USA)using an oligo(dT)18 as the primer according to the manufacturer’s instructions.
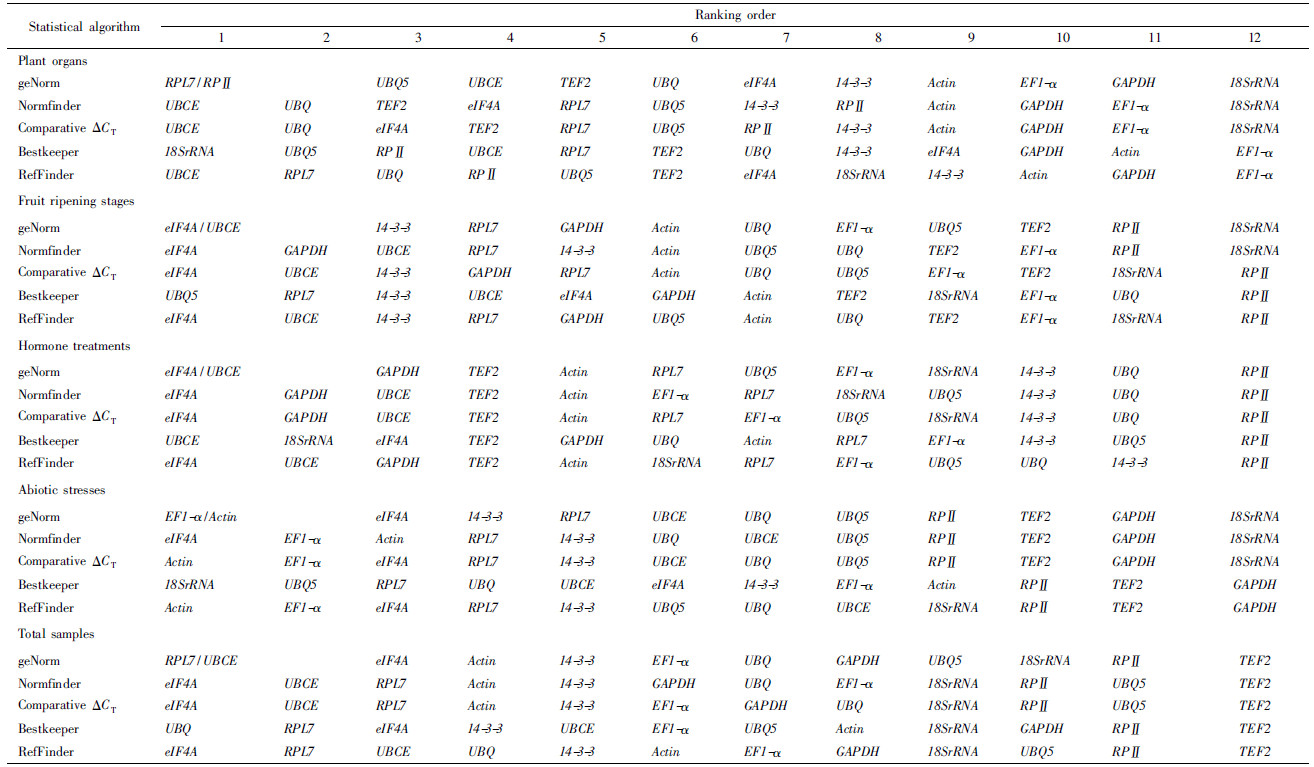
1.3 Primer design and testThe specific primers were used for the amplification of 18SrRNA, Actin and GAPDH. The degenerate primers were designed on the basis of highly conserved coding sequences of other nine tested reference genes(14-3-3, EF1-α, eIF4A, RPⅡ, RPL7, TEF2, UBCE, UBQ and UBQ5)from the National Center for Biotechnology Information(NCBI)database(Tab.1). Primer pairs were designed using Primer Primer V5.0 software(http://www.PremierBiosoft.com/primerdesign/). The PCR conditions were 30 cycles of 15 s at 94 ℃, 30 s at 52 to 60 ℃ depending on the primers(Tab.1), and 90 s at 72 ℃. Purified amplified products of appropriate length were ligated to the pUCm-T vector for sequencing. All reference genes were named based on similarity to known protein sequences with identity ranging from 86% to 99%. Gene sequences were deposited in the GenBank and accession numbers are listed in Tab.2. The specific primers for qRT-PCR were designed based on the obtained sequence above of reference genes using Primer Primer V5.0 with melting temperature(Tm)60 ℃, primer lengths 20-22 bp and amplicon lengths 128-274 bp(Tab.2). The primers for EutPsy gene were from our previous study(Guo et al., 2009). The specificity of all primer pairs was tested by reverse transcription-PCR and also by qRT-PCR, followed by agarose gel electrophoresis and melting curve analysis.
|
|
|
|
qRT-PCR was carried out in 96-well plates with a StepOne PlusTM Real-Time PCR System(Applied Biosystems, Foster, USA)using SYBR® Premix Ex TaqTM Ⅱ Kit(Takara, Toyoto, Japan). PCR reactions were prepared in 20 μL volumes containing2μL of 5-fold diluted cDNA, 10 μL of 2× Power SYBR® Premix Ex TaqTMⅡ, 0.5 μL(10 μmol ·L-1)of each primer, 0.4 μL 50× ROX Reference Dye and 6.6 μL sterile distilled water. Aliquots from the same cDNA solutions were used with all primer sets in each experiment. Negative PCR control with no templates was performed for each primer pair. All reaction mixtures were incubated for 30 s at 95 ℃, followed by 45 cycles of5s at 95 ℃ and 30 s at 60 ℃. The melting curve was obtained by heating the amplicon at 95 ℃ for 15 s followed by a constant increase in the temperature from 60 to 95 ℃, and reading at each 0.3 ℃. For each primer pair, a st and ard curve was generated from 5-fold serial dilutions of the cDNA generated from green fruit. The correlation coefficients(R2) and slope values could be obtained from the st and ard curve, and the corresponding PCR amplification efficiencies(E)were calculated according to the equation E=(10-1/slope-1)×100(Han et al., 2012). The final cycle threshold(CT)values were the mean of at least six values including two or three biological replicates for each treatment and three technical replicates.
1.5 Statistical analyses of gene expression stabilitySeveral algorithms have been developed to study the stability of c and idate reference genes. The geNorm software derives a stability measure(M) and creates a stability ranking via a stepwise exclusion of the least stable gene. It also estimates the number of reference genes necessary for an optimal normalization(Silveira et al., 2009; Zhu et al., 2013). NormFinder software has the advantage of ranking the c and idate reference genes both inter-group and intra-group according to their different expression stability(Han et al., 2012; Zhu et al., 2013). BestKeeper creates an index using the geometric mean of each reference gene’s raw CT values and determines the most stably expressed genes based on coefficient of correlation to the index(Tong et al., 2009; Zhang et al., 2012; Zhu et al., 2013). The comparative ΔCT method evaluates the most stable reference genes by comparing relative expression of “pairs of genes” within each sample, which measures the stability of a gene by the mean of st and ard deviation(SD)values derived from comparison between a reference gene and other c and idate reference genes(Chao et al., 2012; Lu et al., 2013). Based on the rankings from each of the four computational programs aforementioned, a user-friendly web-based comprehensive tool RefFinder(http://www.leonxie.com/referencegene.php?type=reference)assigns an appropriate weight to an individual gene and calculates the geometric mean of their weights for the overall final ranking(Xie et al., 2011; Chao et al., 2012). The lower ranking indicated genes with more stable gene expression.
2 Results 2.1 Verification of primer specificity and PCR amplification efficiencyA total of 12 reference genes were cloned(Tab.1). Gene name, accession number, specific primer sequences for qRT-PCR and amplicon lengths were listed in Tab.2. For each of the potential reference genes, the single peak in qRT-PCR melting curves suggested that each primer pair amplified a unique product. The primer efficiency ranged from 93.56% for UBCE up to 108.22% for 14-3-3 with a correlation coefficient(R2)from 0.990 to 0.998(Tab.2), indicating that the primers worked successfully and gave consistent results.
CT is defined as the number of cycles needed for fluorescence to reach a specific threshold level of detection and is inversely related to the amount of initial RNA template present in the sample. The expression levels of the potential reference genes were determined as CT values. As expected, the median CT values of the genes in the 29 samples were in the range of 16.75 and 30.13 cycles(Fig. 1), showing a high range of variation between them. 18SrRNA was the most abundant transcript, while RPⅡ and UBQ8 were the least abundant transcript with the median CT values about 30 cycles. UBQ, EF1-α, UBCE, Actin, RPL7, GAPDH, 14-3-3, eIF4A and TEF2 displayed median CT values ranging from 19 to 25, which was considered a moderate level of expression. For qRT-PCR normalization, a moderately expressed reference gene is preferred because extremely high or low expression of a reference gene could introduce variability to data analysis.
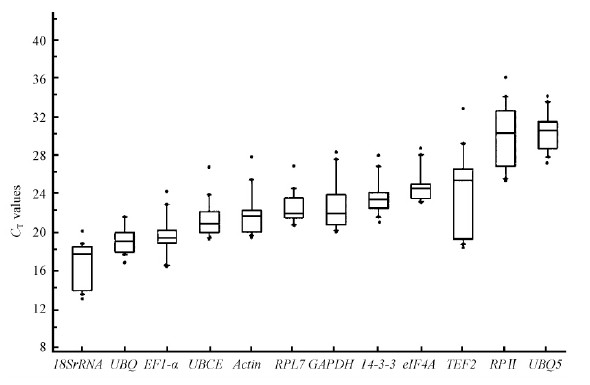 |
Fig.1 Expression levels of candidate reference genes across all samples A line across the box is depicted as the median. The box indicates the 25th and 75th percentiles, whisker caps represent 95% confidence intervals, and black dots represent outliers. |
The stabilities of the 12 potential reference genes were analyzed among different organs(five samples), fruit ripening stages(four samples), hormone treatments(seven samples), and abiotic stresses(13 samples). The CT values for each potential reference gene were used for stability comparison in the NormFinder, geNorm, BestKeeper, and Comparative ΔCT algorithms to identify the best reference genes for qRT-PCR data normalization(Xie et al., 2011; Han et al., 2012; Zhang et al., 2012; Fan et al., 2013; Zhu et al., 2013). Overall ranking of four sets aforementioned was generated using Recommended Comprehensive Ranking method, i.e. RefFinder. Tab.3 summarized the ranking results for 12 potential reference genes. In the meantime, geNorm analysis was also used to determine the optimal number of stable reference genes for accurate normalization(Han et al., 2012). Generally, 0.15 was used as a cutoff value to confirm the optimal number of reference genes(Hong et al., 2008; Han et al., 2012). The evaluation of the organs, fruit-ripening stages or hormone-treated samples did not exhibit any discernible differences in the pairwise variation with the inclusion of a third gene. It is apparent that two stable reference genes would be sufficient for normalizing gene expression for three sets of E. umbellata samples. As for abiotic stress and total samples, the use of three reference genes was recommended although their values were higher than 0.15(Han et al., 2012; Lu et al., 2013).
|
|
In organ samples, six genes(UBCE, RPⅡ, RPL7, 18SrRNA, UBQ and UBQ5)were classified among the top two reference genes in each of four algorithms. The overall ranking of the best reference genes using RefFinder for five organs was UBCE and RPL7. In fruit-ripening samples, five genes(eIF4A, UBCE, UBQ5, GAPDH and RPL7)were identified, and the top two were eIF4A and UBCE based on RefFinder. In hormone treatment set, only eIF4A, UBCE, GAPDH, and 18SrRNA were classified among the top two genes, and the two best were eIF4A and UBCE according to the RefFinder. Likewise, six genes(Actin, eIF4A, EF1-α, 18SrRNA, UBQ5 and RPL7)were identified as the top three reference genes for abiotic stress set, and the three best were Actin, EF1-α, and eIF4A based on the RefFinder.
When including the data obtained from all the 29 samples into the analysis, four genes(eIF4A, RPL7 , UBCE, and UBQ)were identified, of which eIF4A, RPL7 , and UBCE were ranked as the best reference c and idates(Tab.3).
2.3 Reference gene validation for fruit-ripening stagesTo validate the selection of c and idate reference genes, the expression pattern of functional genes EutPsy was inspected using the different reference genes at four ripening stages of E. umbellata fruit(Fig. 2). The top two stable references eIF4A and UBCE in the fruit-ripening stages selected by geNorm, comparative ΔCT and RefFinder, as well as RPL7 selected by BestKeeper were used as internal controls. EutPsy is a key upstream gene for lycopene accumulation during E. umbellata fruit ripening(Guo et al., 2009)similar to tomato(Solanum lycopersicum)Psy-1 gene which is induced by fruit ripening in association with elevated lycopene accumulation(Giuliano et al., 1993). When the three stable reference genes eIF4A, UBCE and RPL7 or the combination of top two eIF4A and UBCE were used for normalization, the trend of the relative transcript abundance of EutPsy were consistent. The expression levels of EutPsy peaked in dark-yellow fruits. However, when the worst reference gene RPⅡ was employed for normalization, EutPsy was expressed in much higher level in yellow fruits that resulted in distinct expression pattern compared to the stable references for normalization(Fig. 2).
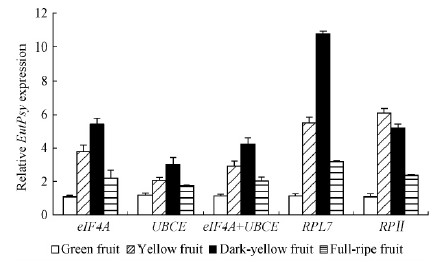 |
Fig.2 Relative quantification of EutPsy expression during fruit ripening using selected reference genes for normalization |
Normalization is one of the key factors affecting the accuracy and reliability of the quantitative gene expression analysis. This study was conducted to identify the appropriate reference genes for gene expression analysis of E. umbellata for studies of different organs, fruit ripening stages, hormone treatments and abiotic stress conditions. To our knowledge, this is the first report on evaluating the expression stability of different potential reference genes for qRT-PCR in the family Elaeagnaceae.
eIF4A protein unwinds RNA secondary structure in the 5′-UTR of mRNAs, which was necessary to allow efficient binding of the small ribosomal subunit and subsequent scanning for the initiator codon. Here, eIF4A was found to rank not only the first from the results of overall ranking of the best reference genes for all samples, but also the first for different fruit ripening stages and hormone treatments, and the third for different abiotic stresses among 12 genes from E. umbellata. The RPL7encoding the transcription regulator and structural constituent of the 60S subunit of the cytosolic ribosome was the second most stable gene in both all samples and five organ samples. The UBCE gene, which is involved in protein degradation through ubiquitination reactions, exhibited the third most stable expression from the results of overall ranking for all samples, also the first one in the different organs and the second one in both different fruit ripening stages and hormone treatments. These genes have been identified as reliable reference genes for transcription normalization in other plants in previous studies. For example, EIF4A showed highly stable expression under most experimental conditions in papaya(Carica papaya)(Zhu et al., 2012), and in the laboratory-grown samples of perennial ryegrass(Lolium perenne)(Lee et al., 2010). E1F4A and UBCE were the best reference genes for various vegetative and reproductive tissues in Brachiaria brizantha(Silveira et al., 2009). UBCE has been noted as displaying superior expression stability and lower absolute expression levels across different nutrient stresses in Arabidopsis thaliana (Czechowski et al., 2005). RPL7 had an equivalent transcript level over five distinct tissue types and was therefore adequate for normalization purposes in Coffea arabica(Barsalobres-Cavallari et al., 2009).
To validate the suitability of the reference genes we identified in this study, the expression profile of EutPsy was assessed in different ripening fruits. During the fruit ripening of E. umbellata, EutPsy is associated with the carotenoid biosynthesis. The data showed that the use of the most stable reference genes UBCE and eI F4 A or the combination of stable references resulted in the trend consistency of the relative transcript abundance of EutPsy. However, the relative abundance displayed a distinct pattern when the most variable reference gene RPⅡ used as an internal control(Fig. 2). Thus, the use of unsuitable references can lead to over- or under-estimation of relative transcript abundance and misinterpretation of data. These results reinforce the importance of validating reference genes prior to experimental applications. The new reference genes will enable more accurate and reliable normalization of qRT-PCR results for gene expression studies in this important plant and other species of the genus Elaeagnus.
| [1] |
Ahmad S D, Sabir S M, Zubair M. 2006. Ecotypes diversity in autumn olive (Elaeagnus umbellata Thunb): A single plant with multiple micronutrient genes. Chemistry and Ecology, 22(6): 509-521.( 3) 3)
|
| [2] |
Barsalobres-Cavallari C F, Severino F E, Maluf M P, et al. 2009. Identification of suitable internal control genes for expression studies in Coffea arabica under different experimental conditions. BMC Molecular Biology, 10: 1.( 4) 4)
|
| [3] |
Chao W S, Do Dgřamaci M, Foley M E, et al. 2012. Selection and validation of endogenous reference genes for qRT-PCR analysis in leafy spurge (Euphorbia esula). PLoS One, 7(8): e42839.( 3) 3)
|
| [4] |
Czechowski T, Stitt M, Altmann T, et al. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology, 139(1): 5-17.( 2) 2)
|
| [5] |
Fan C, Ma J, Guo Q, et al. 2013. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS One, 8(2): e56573.( 2) 2)
|
| [6] |
Fordham I M, Zimmerman R H, Black B L, et al. 2003. Autumn olive: A potential alternative crop. Acta Horticulturae, 626: 437-439.( 4) 4)
|
| [7] |
Giuliano G, Bartley G E, Scolnik P A. 1993. Regulation of carotenoid biosynthesis during tomato development. Plant Cell, 5(4): 379-387.( 1) 1)
|
| [8] |
Guo X L, Yang L, Hu H T, et al. 2009. Cloning and expression analysis of carotenogenic genes during ripening of autumn olive fruit (Elaeagnus umbellata). Journal of Agricultural and Food Chemistry, 57(12): 5334-5339.( 8) 8)
|
| [9] |
Han X, Lu M, Chen Y, et al. 2012. Selection of reliable reference genes for gene expression studies using real-time PCR in tung tree during seed development. PLoS One, 7(8): e43084.( 9) 9)
|
| [10] |
Hong S Y, Seo P J, Yang M S, et al. 2008. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biology, 8: 112.( 5) 5)
|
| [11] |
Huis R, Hawkins S, Neutelings G. 2010. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biology, 10: 71.( 2) 2)
|
| [12] |
Lee J M, Roche J R, Donaghy D J, et al. 2010. Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.). BMC Molecular Biology,11: 8.( 2) 2)
|
| [13] |
Lu Y, Yuan M, Gao X, et al. 2013. Identification and validation of reference genes for gene expression analysis using quantitative PCR in Spodoptera litura (Lepidoptera: Noctuidae). PLoS One, 8(7): e68059.( 3) 3)
|
| [14] |
Nicot N, Hausman J F, Hoffmann L, et al. 2005. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany, 56(421): 2907-2914.( 2) 2)
|
| [15] |
Sabir M S, Ahmad D S, Hussain I M, et al. 2007. Antibacterial activity of Elaeagnus umbellata (Thunb.) a medicinal plant from Pakistan. Saudi Medical Journal, 28(2): 259-263.( 1) 1)
|
| [16] |
Silveira E D, Alves-Ferreira M, Guimarães L A, et al. 2009. Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biology, 9: 84.( 4) 4)
|
| [17] |
Su R L, Hu H T, Wu M, et al. 2013. Isolation and characterisation of a UDP-glucose pyrophosphorylase gene associated with fruit ripening in autumn olive (Elaeagnus umbellata Thunb.). Journal of Horticultural Science & Biotechnology, 88(5): 617-623.( 1) 1)
|
| [18] |
Tong Z, Gao Z, Wang F, et al. 2009. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Molecular Biology, 10: 71.( 4) 4)
|
| [19] |
Xie F, Sun G, Stiller J W, et al. 2011. Genome-wide functional analysis of the cotton transcriptome by creating an integrated EST database. PLoS One, 6(11): e26980.( 2) 2)
|
| [20] |
Zhang Y, Chen D, Smith M A, et al. 2012. Selection of reliable reference genes in Caenorhabditis elegans for analysis of nanotoxicity. PLoS One, 7(3): e31849.( 2) 2)
|
| [21] |
Zhu J, Zhang L, Li W, et al. 2013. Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PLoS One, 8(1): e53196.( 5) 5)
|
| [22] |
Zhu X, Li X, Chen W, et al. 2012. Evaluation of new reference genes in papaya for accurate transcript normalization under different experimental conditions. PLoS One, 7(8): e44405.( 2) 2)
|
 2015, Vol. 51
2015, Vol. 51