文章信息
- 陈昊, 谭晓风
- Chen Hao, Tan Xiaofeng
- 基于油脂合成期油桐种仁转录组数据的α-亚麻酸代谢途径解析
- Identification of α-Linolenic Acid Metabolism Pathway Based on Transcriptome Data of Vernicia fordii Kernels during Tung Oil Synthesis Stage
- 林业科学, 2015, 51(3): 41-48
- Scientia Silvae Sinicae, 2015, 51(3): 41-48.
- DOI: 10.11707/j.1001-7488.20150306
-
文章历史
- 收稿日期:2014-07-21
- 修回日期:2014-08-22
-
作者相关文章
油桐(Vernicia fordii)是我国四大木本油料植物之一,其种仁中榨出的桐油的经济利用价值极高,能够用于优质油漆、油墨等工业原料的制造,发展桐油产业对于发展我国新材料工业、缓解我国能源危机具有重要的战略意义(黄坤等,2008;杨颖等,2010;谭晓风等,2011)。桐油中脂肪酸的主要成分是α-桐酸(α-eleostearic acid),其含量占桐油脂肪酸总量的70%(w/w)以上(Bickford et al.,1953;傅伟昌等,2008;刘金龙等,2011)。目前,α-桐酸的形成机理还不明确,致使只能采用常规的杂交和选择育种,不能有效地从分子水平对油桐进行遗传改良,这严重阻碍了桐油产业的发展。
所有共轭十八碳三烯酸被总称为共轭亚麻酸(conjugated linolenic acid,CLNA),因此桐酸是CLNA中的一个亚类。桐油中的桐酸又可分为α-桐酸(C18:3Δ9cis,11trans,13trans)和β-桐酸(C18: 3Δ9cis,11trans,13trans)2类,其中绝大部分为α-桐酸(Bickford et al.,1953;Greenfield,1959;傅伟昌等,2008;刘金龙等,2011)。高α-桐酸含量导致桐油的不饱和程度很高,从而使得桐油成为一种优质的干性油,被广泛应用于工业生产(Wang et al.,2000;蒲侠等,2003;Li et al.,2003;黄坤等,2008)。由于α-桐酸具有极高的应用价值,有学者尝试通过化学合成和基因工程的方法生产α-桐酸,但这些方法都存在步骤复杂、产率低、不能进行大规模生产等缺陷,因而从天然植物资源中分离仍是目前最行之有效的获取α-桐酸的途径(Cahoon et al.,1999;2006)。有鉴于此,育种家们希望尽可能地通过遗传改良来提高单位面积的桐油产量或增加单位质量桐油中α-桐酸的含量。解析油桐α-桐酸的形成机理将为油桐的遗传改良提供理论指导。
与拟南芥(Arabidopsis thaliana)、水稻(Oryza sativa)和毛果杨(Populus trichocarpa)等传统模式植物相比,油桐还没有进行全基因组测序,这对油桐功能基因的克隆和功能研究造成了一定的困难。转录组测序(RNA-Seq)这一转录组学研究方法基于第二代测序平台进行,它比传统的基因芯片数据通量更高、成本更低、灵敏度更高、重复性更好,而且不需知道待测物种的基因序列,已成为当今转录组学研究的主要方法(Marioni et al.,2008;O’Loughlin et al.,2012;Shao et al.,2012;Torales et al.,2012;Yang et al.,2012)。因此,新一代测序技术的发展为解决这一难题提供了必要的技术手段。尽管如此,植物体内α-桐酸代谢通路的相关研究还未见报道,因此哪些酶基因参与了α-桐酸的代谢仍不清楚,这对通过序列同源比对的方法直接筛选油桐α-桐酸代谢通路相关酶基因造成了一定的困难。作为α-桐酸的同分异构体,α-亚麻酸(C18:3Δ9cis,12cis,15cis)与α-桐酸仅存在不饱和双键位置的差异,这意味着二者的代谢通路有很大程度的重叠。参与α-亚麻酸代谢的酶基因已有研究,鉴于α-桐酸和α-亚麻酸分子结构的高度相似性,这些基因很可能也参与了α-桐酸的代谢。通过序列的同源性比对,找到油桐α-亚麻酸代谢相关基因,随后通过分析这些基因的表达模式及功能,确定其与α-桐酸代谢的相关性,将为油桐α-桐酸代谢通路的解析提供突破口。另外,作者对36个油桐家系的研究发现,成熟的油桐种仁中,α-桐酸和α-亚麻酸的含量均值分别为77.15%和0.76%,α-桐酸的含量远大于α-亚麻酸的含量。油桐种仁在发育过程中油脂大量积累,但在这一过程中究竟是直接生成α-桐酸还是先生成α-亚麻酸后再将其转化为α-桐酸这一问题的解析同样有赖于对α-亚麻酸代谢通路的研究。然而油桐的全基因组序列还没有被测定,无可用参考信息,因此,本研究基于利用RNA-Seq技术获取的油桐种仁转录组的测序数据,通过与其他物种的序列进行同源性比对和代谢通路富集性分析,解析了油桐α-亚麻酸代谢通路并分析了通路中关键酶基因在油脂合成期的表达模式。研究结果能为在此基础上进行的后续油桐α-桐酸代谢通路解析提供理论突破口。此外,通过调控这些基因的表达模式以及开发与之紧密连锁的分子标记,可大大加快油桐遗传改良和分子育种的进程。
1 材料与方法 1.1 试验材料以油桐代表性地方品种‘泸溪葡萄桐’(Vernicia fordii ‘Putaotong’)6月(Ⅰ期)、8月(Ⅱ期)和10月(Ⅲ期)采收的种仁为材料,提取种仁RNA进行RNA-Seq测序。前人的研究表明,油桐种仁中油脂的合成可分为起始期(6月)、快速积累期(8月)和停滞期(10月)3个阶段(蔺定运等,1980;王汉涛等,1985)。
1.2 总RNA提取与测序文库构建使用Invitrogen公司的Trizol试剂进行总RNA的提取。进行RNA-Seq测序的总RNA在使用前需经毛细管电泳和Agilent 2100 Bioanalyzer仪器分析其完整度(1.8<OD260/OD280< 2.2,28S :18S>1.0,RIN值≥6.5),用分光光度计检测其浓度(浓度≥400 ng ·μL-1,总量≥20 μg)。建库所用第1链cDNA的合成使用SuperScriptTM II RT试剂盒(Invitrogen)进行,随后以置换法合成双链cDNA。在合成双链cDNA的基础上按照Illumina公司提供的操作手册进行双末端测序文库的构建,构建好的测序文库置于高通量测序平台HiSeq 2000上测序。
1.3 测序数据的处理通过RNA-Seq获取的原始数据(raw reads)的实质是众多短核苷酸序列组成的集合。这些原始数据需经低质量序列(N的比例大于5%,20%以上为Q≤10的序列)和接头(adaptor)序列的剔除才能获得可用于后续分析的数据(clean reads)。随后将首尾重叠的reads序列通过Trinity(Grabherr et al.,2011)这一主流序列组装软件组装成序列两端不能再延伸的Unigene序列。最后将Unigene序列在蛋白质数据库(non-redundant,nr;Swiss-Protein,Swiss-Prot;Kyoto Encyclopedia of Genes and Genomes,KEGG;Cluster of Orthologous Groups of Proteins,COG)中与其他物种的同源序列进行比对,从而确定Unigene序列的序列方向。
1.4 Unigene的功能注释和差异表达分析将Unigene序列在1.3所述的蛋白质数据库中进行比对以获得其蛋白质的功能注释。随后分别利用软件Blast2GO(Conesa et al.,2005)和KEGG数据库进行Unigene序列的GO(Gene Ontology)分类和代谢途径(Pathway)富集性分析。采用FPKM(fragments per kb per million fragments)算法(Mortazavi et al.,2008)从转录组数据中获取Unigene的表达丰度从而筛选在不同油脂合成期之间差异表达的Unigene。同一Unigene的表达丰度在2个不同时期间进行比较时,如Unigene的FDR值≤0.001且测序丰度的比值在进行log2转化后所得的|log2Ratio|≥1,则认为其为一个差异表达Unigene(Audic et al.,1997)。随机抽取转录组测序数据,使用Sanger测序和real-time PCR验证转录组测序数据的准确性。
2 结果与分析 2.1 RNA质量检测油桐种仁中含有大量的油脂,这对RNA的质量会造成影响,因此提取的总RNA在用于构建测序文库前需进行质量评估。通过毛细管电泳和Agilent 2100 Bioanalyzer仪器分析油桐种仁总RNA的完整度和浓度,发现各样本RNA没有降解(图 1),其完整度(RIN>7.0,28S :18S>1.5)和浓度(≥400 ng ·μL-1)均符合测序要求。
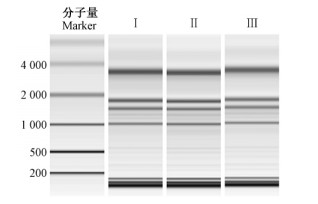 |
图 1 油桐种仁3个不同发育时期总RNA的毛细管电泳 Fig. 1 Capillary electrophoresis diagram of total RNA of V. fordii kernels in three different developmental stages Ⅰ,Ⅱ和Ⅲ分别表示油桐种仁的3个发育时期。 Ⅰ, Ⅱ and Ⅲ represent three developmental stages of V. fordii kernel, respectively. |
通过对Ⅰ-Ⅲ期的油桐种仁RNA进行测序,共获得58 439条长度为200~3 000个核苷酸的非冗余Unigene序列,其中Ⅰ,Ⅱ和Ⅲ期分别获得了61 001,54679和44 495条Unigene序列。共有41 059条Unigene序列能够与公共数据库中的已知基因匹配,占所有非冗余基因的70.3%(表 1)。不同长度的非冗余Unigene序列与数据库中序列匹配的效率不同,越长的序列匹配效率越高(图 2)。序列长度大于2 000 bp的序列匹配效率达到了98.28%,而500~1 000 bp和100~500bp的序列分别只有78.86%和48.99%的匹配效率。
|
|
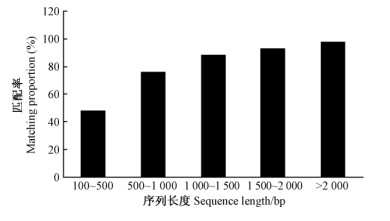 |
图 2 不同碱基对长度的Unigene序列在GenBank数据库中的匹配效率 Fig. 2 Matching percentage of Unigene sequences with different lengths of base pairs to entries in the GenBank database |
Pathway富集性分析表明,3个种仁油脂合成期的转录组数据中共有105个Unigene可被富集于α-亚麻酸代谢(linolenic acid metabolism)途径,占所有非冗余Unigene的0.47%。从3个转录组数据的两两比较中分别鉴别出一些差异表达Unigene,其中也有一些可被富集于α-亚麻酸代谢途径(图 3)。通过在KEGG数据库中进行检索后发现,105个Unigene序列分别对应于14个α-亚麻酸代谢途径关键酶基因(表 2),这些基因在其他物种中都有同源基因与之对应。随后利用转录组数据对这些基因在油桐种仁3个不同油脂合成期的表达变化规律进行解析(表 2)。随机挑选其中4个基因,利用real-time PCR检测其油脂合成期的表达模式,结果与测序数据一致,表明测序数据准确可靠(图 4)。
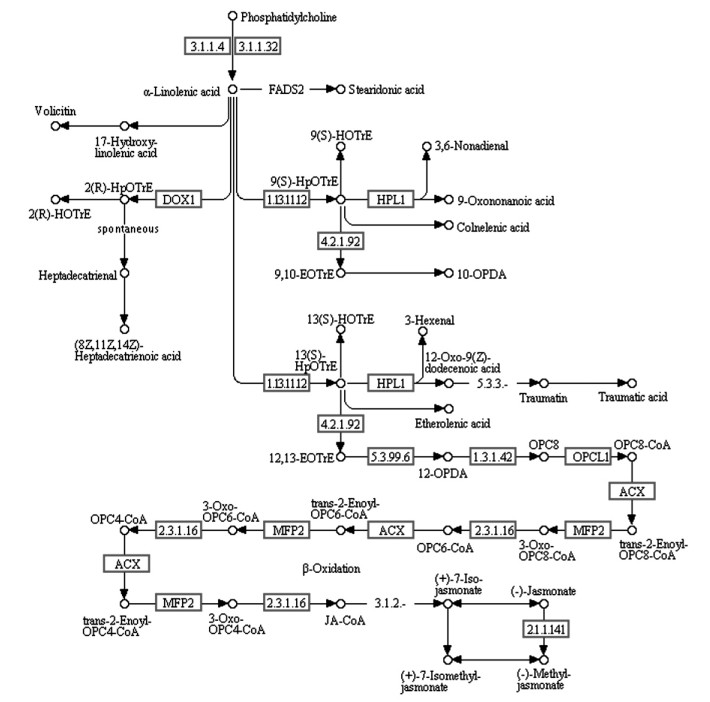 |
图 3 RNA-Seq检测到的油桐油脂合成期α-亚麻酸代谢途径差异表达基因
Fig. 3 Differentially expressed genes in α-linolenic acid metabolism pathway detected by RAN-Seq during tung oil synthesis stage of V. fordii
矩形框标示油桐种仁转录组两两比较时的差异表达基因。代谢途径图引自KEGG数据库(登录号:map00592)。 Rectangular boxes indicate differentially expressed genes across pairwise comparisons of V. fordii kernels transcriptome. Metabolic pathway map was cited from KEGG database (accession number: map00592). |
|
|
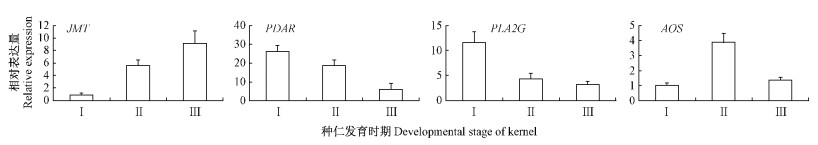 |
图 4 油桐油脂合成期部分α-亚麻酸代谢途径基因表达模式的real-time PCR检测 Fig. 4 Expression profiles of partial genes involved in α-linolenic acid metabolism pathway detected by real-time PCR during tung oil synthesis stage of V. fordii Ⅰ,Ⅱ和Ⅲ分别表示油桐种仁的3个发育时期。 Ⅰ, Ⅱ and Ⅲ represent three developmental stages of V. fordii kernel, respectively. |
通过基因表达模式分析发现,整体上与合成代谢相关的基因在油脂合成期呈现上调的表达模式,而与分解代谢相关的基因则呈现下调的表达模式。例如,与α-亚麻酸合成代谢相关的乙酰辅酶A连接酶(OPCL1)基因和乙酰辅酶A酰基转移酶(ACAA1)基因在油脂合成期持续上调表达(表 2)。与α-亚麻酸分解代谢相关的酰基辅酶A氧化酶(ACX)和烯酰辅酶A水合酶(MFP2)分别催化脂肪酸β-氧化的第1和第2步反应,生成乙酰辅酶A和能量 (Bahnson et al.,2002;Carrozzo et al.,2008)。ACX和MFP2的表达量在油脂合成期持续下调,这一表达模式降低了种仁中ACX和MFP2的含量,从而有利于油脂合成期脂肪酸的大量积累(表 2)。磷脂酶(Phospholipase,PL)是催化生物体内磷脂分解的重要酶类,细胞中膜结构的损伤修复、信号转导等生理过程都离不开磷脂酶的参与(Casado et al.,2012)。在油脂合成期内,油桐α-亚麻酸代谢途径中的2个磷脂酶基因pldA和PLA2G的表达量始终保持下降的趋势(表 2)。
3 结论与讨论油桐的全基因测序还未完成,可用的核酸序列信息过少,这严重制约了油桐分子生物学研究的发展,从而使油桐重要经济性状分子机理的解析变得困难。RNA-Seq这一基于第二代测序技术的转录组研究方法的优越性在于其在进行转录组研究时会对cDNA进行测序,因此无需知道被研物种的序列信息。另外,这一方法与传统的转录组研究手段(cDNA微阵列,LongSAGE,MPSS)相比,灵敏度大大提高,甚至可检测到细胞中只有几个拷贝的转录本(Alagna et al.,2009;González-Ballester et al.,2010;Zenoni et al.,2010;Bennetzen et al.,2012)。本研究在获得油桐种仁油脂合成期转录组数据的基础上,通过一系列的生物信息学分析,揭示了油桐的α-亚麻酸代谢途径,并分析了途径中基因的表达规律,这为进一步研究油桐α-亚麻酸代谢途径中关键酶基因的功能提供了线索,也为油桐α-桐酸代谢通路的解析提供了必要的数据支持。
转录组测序数据中共有41 059条Unigene序列能够与公共数据库中的已知基因匹配,占所有非冗余基因的70.3%(表 1), 这意味着有30%的Unigene在公共数据库中没有检测到同源序列。前人的研究表明,大约有64%通过RNA-Seq从人转录组中获得的表达序列标签(Expressed sequence tag,EST)能够较好地比对到已经注释过的人类基因上(Mane et al.,2009)。其他植物的转录组研究中同样有比例不等(13%~80%)的序列不能进行有效的注释,这些序列所占的比例受到物种、测序深度和BLAST程序参数等因素的影响(Wang et al.,2010;Blanca et al.,2011;Ness et al.,2011)。考虑到油桐缺少有用的基因组序列信息,本研究对Unigene的注释率符合转录组测序数据注释标准。
α-亚麻酸是生物体脂肪酸的主要成分之一,也是α-桐酸的同分异构体,因此对α-亚麻酸代谢途径基因的研究有助于阐述α-桐酸形成的分子机理。植物的α-亚麻酸代谢是一个多种酶类共同参与的复杂过程,任何一种酶类的表达变化都会对α-亚麻酸代谢造成影响,这也是不同植物乃至同一植物的不同组织器官中α-亚麻酸含量具有显著差异的分子遗传基础。因此,通过对α-亚麻酸代谢途径基因表达模式的整体阐述,有助于更深入地揭示油桐α-亚麻酸代谢这一生理过程的分子机制。在油桐α-亚麻酸代谢通路中,与合成代谢相关的基因,如乙酰辅酶A连接酶(OPCL1)基因和乙酰辅酶A酰基转移酶(ACAA1)基因在油脂合成期持续上调表达(表 2),与分解代谢相关的基因,如酰基辅酶A氧化酶(ACX)基因和烯酰辅酶A水合酶(MFP2)基因则持续下调表达。磷脂分解代谢的关键酶是磷脂酶(Phospholipase,PL),细胞中膜结构的损伤修复、信号转导等生理过程都离不开磷脂酶的参与(Casado et al.,2012)。在油脂合成期内,油桐α-亚麻酸代谢途径中的2个磷脂酶基因pldA和PLA2G持续下调表达(表 2)。从整体上看,油桐α-亚麻酸代谢通路中与合成代谢相关的基因呈现上调表达的趋势,而与分解代谢相关的基因则表现出下调趋势,这与油桐油脂合成期油脂的大量积累这一表型相吻合。
第二代测序技术能够获得大量的油桐转录本序列,这将为后续的研究带来极大的便利。尽管利用第二代测序技术研究转录组时技术优势明显,但也存在一些问题。首先,随着第二代测序技术的发展,如何高效地从越来越多的数据中发掘相关信息,将对生物信息学的发展提出挑战(van Vliet,2010);其次,高通量测序的成本仍然偏高,尤其在小规模测序,如质粒测序、PCR产物测序等方面,没有成本优势;第三,高通量测序所需的起始样本量大,这使其应用受到了限制。尽管第二代测序技术存在上述缺陷,但随着技术水平和生物信息学分析方法的不断发展,第二代测序技术的测序成本必将逐步降低,从而使其得到更广泛的应用。
| [1] |
傅伟昌,顾小红,陶冠军,等.2008.桐油脂肪酸组成分析和甘三酯结构判定.天然产物研究与开发,(6): 964-968. (Fu W C,Gu X H,Tao G J,et al. 2008.Analysis of fatty acids and the structure identification of triacylglycerols in tung oil.Natural Product Research and Development,(8): 964-968[in Chinese]).(  2) 2)
|
| [2] |
黄坤,夏建陵.2008.桐油及其衍生物的改性在高分子材料中的应用进展.化工进展,27(10): 1588-1592. (Huang K,Xia J L.2008.Progress of modification of tung oil and its derivatives in the application of polymer materials.Chemical Industry and Engineering Progress,27(10): 1588-1592[in Chinese]).(  2) 2)
|
| [3] |
蔺定运,倪善庆.1980.油桐枝、叶和果实物质代谢的初步研究.植物生理学通讯,(3): 37-40. (Lin D Y,Ni S Q.1980.Preliminary researches on material metabolism of branches,leaves and fruits of tung tree.Plant Physiology Communication,(3): 37-40[in Chinese]).(  1) 1)
|
| [4] |
刘金龙,郑小江,郑威,等.2011.油桐品种五爪桐含油量及桐油质量研究.湖北农业科学,50(10): 2031-2035. (Liu J L,Zheng X J,Zheng W,et al. 2011.Study on the oil content and quality of Vernicia fordii cv.five fingernail tung.Hubei Agricultural Sciences,50(10): 2031-2035[in Chinese]).(  2) 2)
|
| [5] |
蒲侠,张兴华,童速玲,等.2003.桐油改性的研究进展及应用前景.林产化工通讯,37(6): 41-46. (Pu X,Zhang X H,Tong S L,et al. 2003.Study and application prospect of modified tung oil.Journal of Chemical Industry of Forest Products,37(6): 41-46[in Chinese]).(  1) 1)
|
| [6] |
谭晓风,蒋桂雄,谭方有,等.2011.我国油桐产业化发展战略调查研究报告.经济林研究,29(3): 1-7. (Tan X F,Jiang G X,Tan F Y,et al. 2011.Research report on industrialization development strategy of Vernicia fordii in China.Nonwood Forest Research,29(3): 1-7[in Chinese]).(  1) 1)
|
| [7] |
王汉涛,段聪仁,徐树华,等.1985.油桐种仁与油脂形成规律的研究.经济林研究,3(2): 29-35. (Wang H T,Duan C R,Xu S H,et al. 1985.Researches on the law of the formulation of seeds and their oil in tung-oil trees.Nonwood Forest Research,3(2): 29-35[in Chinese]).(  1) 1)
|
| [8] |
杨颖,田从学.2010.我国生物柴油产业现状及发展对策.中国粮油学报,25(2): 150-154. (Yang Y,Tian C X.2010.Current status and development strategies of biodiesel industry in China.Journal of the Chinese Cereals and Oils Association,25(2): 150-154[in Chinese]).(  1) 1)
|
| [9] |
Alagna F,D'Agostino N,Torchia L,et al. 2009.Comparative 454 pyrosequencing of transcripts from two olive genotypes during fruit development.BMC Genomics,10: 399.( 1) 1)
|
| [10] |
Audic S,Claverie J M.1997.The significance of digital gene expression profiles.Genome Res,7(10): 986-995.( 1) 1)
|
| [11] |
Bahnson B J,Anderson V E,Petsko G A.2002.Structural mechanism of enoyl-CoA hydratase: three atoms from a single water are added in either an E1cb stepwise or concerted fashion.Biochemistry,41(8): 2621-2629.( 1) 1)
|
| [12] |
Bennetzen J L,Schmutz J,Wang H,et al. 2012.Reference genome sequence of the model plant Setaria.Nat Biotechnol,30(6): 555-561.( 1) 1)
|
| [13] |
Bickford W G,DuPréE F,Mack C H,et al. 1953.The infrared spectra and the structural relationships between alpha-and beta-eleostearic acids and their maleic anhydride adducts.J Am Oil Chem Soc,30(9): 376-381.( 2) 2)
|
| [14] |
Blanca J,Cañizares J,Roig C,et al. 2011.Transcriptome characterization and high throughput SSRs and SNPs discovery in Cucurbita pepo(Cucurbitaceae).BMC Genomics,12: 104.( 1) 1)
|
| [15] |
Cahoon E B,Carlson T J,Ripp K G,et al. 1999.Biosynthetic origin of conjugated double bonds: production of fatty acid components of high-value drying oils in transgenic soybean embryos.Proc Natl Acad Sci USA,96(22): 12935-12940.( 1) 1)
|
| [16] |
Cahoon E B,Dietrich C R,Meyer K,et al. 2006.Conjugated fatty acids accumulate to high levels in phospholipids of metabolically engineered soybean and Arabidopsis seeds.Phytochemistry,67(12): 1166-1176.( 1) 1)
|
| [17] |
Carrozzo R,Bellini C,Lucioli S,et al. 2008.Peroxisomal acyl-CoA-oxidase deficiency: two new cases.Am J Med Genet A,146(13): 1676-1681.( 1) 1)
|
| [18] |
Casado V,Martín D,Torres C,et al. 2012.Phospholipases in food industry: a review.Methods Mol Biol,861: 495-523.( 2) 2)
|
| [19] |
Conesa A,Götz S,García-Gómez J M,et al. 2005.Blast2GO: a universal tool for annotation,visualization and analysis in functional genomics research.Bioinformatics,21(18): 3674-3676.( 1) 1)
|
| [20] |
González-Ballester D,Casero D,Cokus S,et al. 2010.RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival.Plant Cell,22(6): 2058-2084.( 1) 1)
|
| [21] |
Grabherr M G,Haas B J,Yassour M,et al. 2011.Full-length transcriptome assembly from RNA-Seq data without a reference genome.Nat Biotechnol,29(7): 644-652.( 1) 1)
|
| [22] |
Greenfield J.1959.Tung oil.J Am Oil Chem Soc,36(11): 565-574.( 1) 1)
|
| [23] |
Li F,Larock R C.2003.Synthesis,structure and properties of new tung oil-styrene-divinylbenzene copolymers prepared by thermal polymerization.Biomacromolecules,4(4): 1018-1025.( 1) 1)
|
| [24] |
Mane S P,Evans C,Cooper K L,et al. 2009.Transcriptome sequencing of the Microarray Quality Control(MAQC)RNA reference samples using next generation sequencing.BMC Genomics,10: 264.( 1) 1)
|
| [25] |
Marioni J C,Mason C E,Mane S M,et al. 2008.RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays.Genome Res,18(9): 1509-1517.( 1) 1)
|
| [26] |
Mortazavi A,Williams B A,McCue K,et al. 2008.Mapping and quantifying mammalian transcriptomes by RNA-Seq.Nat Methods,5(7): 621-628.( 1) 1)
|
| [27] |
Ness R W,Siol M,Barrett S C.2011.De novo sequence assembly and characterization of the floral transcriptome in cross-and self-fertilizing plants.BMC Genomics,12: 298.( 1) 1)
|
| [28] |
O'Loughlin A,Lynn D J,McGee M,et al. 2012.Transcriptomic analysis of the stress response to weaning at housing in bovine leukocytes using RNA-seq technology.BMC Genomics,13: 250.( 1) 1)
|
| [29] |
Shao W,Zhao Q Y,Wang X Y,et al. 2012.Alternative splicing and trans-splicing events revealed by analysis of the Bombyx mori transcriptome.RNA,18(7): 1395-1407.( 1) 1)
|
| [30] |
Torales S L,Rivarola M,Pomponio M F,et al. 2012.Transcriptome survey of Patagonian southern beech Nothofagus nervosa(=N.alpina): assembly,annotation and molecular marker discovery.BMC Genomics,13: 291.( 1) 1)
|
| [31] |
van Vliet A H.2010.Next generation sequencing of microbial transcriptomes: challenges and opportunities.FEMS Microbiol Lett,302(1): 1-7.( 1) 1)
|
| [32] |
Wang C,Jones F N.2000.Stability and film properties of tung oil modified soybean alkyd emulsion.J Appl Polym Sci,78(9): 1698-1706.( 1) 1)
|
| [33] |
Wang X W,Luan J B,Li J M,et al. 2010.De novo characterization of a whitefly transcriptome and analysis of its gene expression during development.BMC Genomics,11: 400.( 1) 1)
|
| [34] |
Yang D,Liu Q,Yang M,et al. 2012.RNA-seq liver transcriptome analysis reveals an activated MHC-I pathway and an inhibited MHC-II pathway at the early stage of vaccine immunization in zebrafish.BMC Genomics,13: 319.( 1) 1)
|
| [35] |
Zenoni S,Ferrarini A,Giacomelli E,et al. 2010.Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq.Plant Physiol,152(4): 1787-1795.( 1) 1)
|
 2015, Vol. 51
2015, Vol. 51

