文章信息
- 张敏, 周鹏, 季永华
- Zhang Min, Zhou Peng, Ji Yonghua
- 苦楝-小麦农林复合生态系统土壤真菌群落结构分析
- Analysis on the Soil Fungal Community Structure in Melia azedarach- Triticum aestivum Agroforestry Ecosystem
- 林业科学, 2015, 51(10): 26-34
- Scientia Silvae Sinicae, 2015, 51(10): 26-34.
- DOI: 10.11707/j.1001-7488.20151004
-
文章历史
- 收稿日期:2014-12-17
- 修回日期:2015-04-06
-
作者相关文章
2. 南京林业大学生物与环境学院 南京 210037
2. College of Biology and the Environment, Nanjing Forestry University Nanjing 210037
根际是指土壤中紧邻植物根部的一个狭窄区域,这一区域直接受到植物根系活动及土壤微生物的影响(Hinsinger et al., 2009)。植物根际形成一个相对稳定的微环境并且由根系分泌物提供了丰富的营养物质,从而吸引大量微生物定居(Saito et al., 2007)。研究表明,宿主植物可以通过根系分泌物调控土壤微生物的群落结构(Haichar et al., 2008),土壤类型也会影响根际微生物群落(Berg et al., 2009),但究竟哪个因素更为重要目前尚无定论。根际微生物,特别是真菌会对宿主植物的生长产生重要影响。如菌根真菌可以促进根系对水分和矿质元素的吸收、改善土壤的物理性质;某些真菌可以分泌抗菌物质抑制土壤病原菌的生长,从而保护宿主植物免受病原菌侵袭;内生真菌有助于协助宿主植物应对不良环境胁迫;土壤真菌还能分解落叶等有机物为植物提供营养物质(Finlay,2008; Mendes et al., 2013; Berendsen et al., 2012; Khan et al., 2012)等。土壤真菌对植物生长也会产生不利影响,如某些真菌可能引发植物病害、影响作物产量甚至导致植物死亡(Mendes et al., 2013)。此外,宿主植物也会对根际真菌的生长产生影响,如植物凋落物可以为根际真菌提供碳源、植物根系分泌物可以选择性抑制某些真菌而促进另外一些真菌的生长(Urbanová et al.,2015)。
农林复合生态系统中的木本植物可以改善土壤的理化性质,从而影响土壤微生物群落的结构与功能(Fanish et al., 2013; Vallejo et al., 2012; Rivest et al., 2013)。然而,目前有关农林复合生态系统中微生物多样性和群落结构的研究并不多见。对根际真菌多样性及其群落结构的研究将有助于深入了解其生态功能。由于许多真菌在实验室条件下不能培养,因此传统的培养方法往往会低估土壤真菌菌群多样性。此外,培养方法费时费力也限制了其在真菌群落分析中的应用。近年来,为克服传统培养方法的缺陷,真菌生态学家开发出一系列基于PCR技术的分析方法,如变性梯度凝胶电泳(DGGE)、末端限制片段长度多态性(T-RFLP)、单链构象多态性(SSCP)等。这些方法中的PCR-DGGE技术已被广泛应用于微生物多样性研究并已取得重要进展(Broeckling et al., 2008; Ketema et al., 2014; Manici et al., 2010)。本研究采用18S rDNA PCR-DGGE及克隆技术分析了苦楝 (Melia azedarach) -小麦 (Triticum aestivum)农林复合生态系统根际及非根际土壤真菌群落结构,以期揭示农林复合生态系统中伴生树种对作物根际真菌群落的影响,为农林复合经营实践提供一定的理论参考。
1 材料与方法 1.1 试验点自然概况试验点设在江苏射阳林场(33°33′30″—33°37′30″ N,120°24′35″—120°30′35″ E)。该林场位于江苏中部沿海的淤泥质海岸线上,面积约2 000 hm2,其中林地1 500 hm2,疏林及灌木林地100 hm2,森林覆盖率达80%以上。射阳林场属于北亚热带海洋性季风湿润气候,年平均气温14.5 ℃,1月平均最低气温8.0 ℃,7月平均最高气温27.1 ℃,平均气温≥10 ℃的平均日数为221天。年均降水量为1 069.0 mm,年均蒸发量为1 407.4 mm。年均日照时数为2 218.8 h。地下水位一般为1.5~2.0 m,水质多为弱矿水,pH值8.0。
1.2 试验设计本研究农林复合经营模式为苦楝-小麦间作,并以单作小麦作为对照。苦楝-小麦间作田和纯小麦田于2010年建立。苦楝株行距为3 m×4 m,小麦栽培时距离树行两侧各1 m。单作和间作的生产管理方式相同。
1.3 土壤样品采集2012年5月,分别在苦楝-小麦间作田和纯小麦田中建立标准样地,大小均为1.3 hm2。为了减少土壤空间异质性带来的误差,每个样地内设定3个2 m×2 m的小样方,各样方间相距50 m。根际与非根际土壤均采用5点混合采样法。根际土壤采样时,挖取距地表10~15 cm细根(直径<5 mm),保留距根表4 mm左右的细粒土壤 ;非根际土壤采样距地表10~15 cm,过80目筛。将采集的细根和非根际土壤装入无菌聚乙烯塑料袋中,编号后放入低温样品采集保温盒中带回实验室,放入-80 ℃冰箱保存备用。根际土壤采用洗涤法获得。
1.4 土壤DNA提取及PCR扩增土壤总DNA的提取采用Omega公司的E.Z.N.A.TM Soil DNA Kit试剂盒,提取方法参照试剂盒说明进行操作。为获得根际真菌DNA,将5 g细根放入50 mL 0.85%的无菌NaCl溶液中振荡30 min,弃去细根后1 000 r·min-1离心10 min,所得沉淀用于提取根际真菌DNA(Teixeira et al., 2010)。提取的DNA存放于-80 ℃冰箱待用。真菌群落多样性评估按照Vainio等(2000)报道的方法,采用通用引物GC-FR1(5′-CCC CCG CCG CGC GCG GCG GGC GGG GCGGGG GCA CGG GCC GAI CCA TTC AAT CGG TAI T-3′)和FF390(5′-CGA TAA CGA ACG AGA CCT-3′)扩增18S rDNA基因目的片段,其中引物FR1的5′末端添加40 bp的GC序列。PCR采用50 μL反应体系,扩增条件:95 ℃ 8 min;95 ℃ 30 s,50 ℃ 45 s,72 ℃ 2 min,30次循环;72 ℃延伸10 min。PCR产物先用Omega公司的E.Z.N.A. Cycle-Pure Kit试剂盒纯化,再进行DGGE分析。
1.5 DGGE分析变性梯度凝胶电泳(DGGE)采用CBS公司的DGGE-2001突变检测系统进行,使用8%的聚丙烯酰胺凝胶,变性剂浓度范围为40%~60%(100%变性剂指40%去离子甲酰胺+7 mol·L-1尿素)。电泳条件:1×TAE电泳缓冲液,先用20 V恒压电泳15 min,再用100 V恒压、60 ℃恒温电泳16 h。电泳结束后,凝胶用SYBR Green I(1:10 000稀释)染色30 min。染色后的凝胶用Bio-Rad公司的ChemiDOC XRS凝胶成像系统拍照。
DGGE条带图谱用Quantity One 1-D软件分析,不同处理间微生物群落结构的相似性采用非加权组算术平均法(unweighted pair-group method with arithmetic means,UPGMA)分析比较。各样品真菌多样性采用物种丰富度(species richness,S)、香侬(Shannon-Wiener)多样性指数(H)和均匀度指数(equitability index,E)3个指标评价。香侬多样性指数和均匀度指数的计算公式如下:H=-∑PilnPi; E= H/lnS。其中,Pi=ni/N,ni是第i条带的强度,N为样品中所有条带的总强度(Yu et al., 2004),S为某个样品中的条带总数。主成分分析参照Ogino等(2001)的方法采用Canoco软件。
1.6 DGGE 条带序列测定及系统发育树构建在紫外灯下将DGGE凝胶上的条带切下后放入盛有E.Z.N.A.TM Poly-Gel DNA(Omega公司生产)提取试剂的无菌EP管中,按试剂操作说明纯化凝胶中的DNA片段。纯化的DNA片段再次用FF390/FR1-GC引物对进行PCR扩增,之后再次进行DGGE凝胶电泳(变性剂浓度40%~60%)以确定所切片段的纯度。再次割胶并用不含GC序列的FF390/FR1引物对进行PCR扩增。将PCR产物与pEASY-T1 Simple Cloning载体(北京全式金公司)连接后转化大肠杆菌 (Eescheria coli)感受态细胞。采用M13F和M13R引物进行PCR扩增,鉴定阳性重组子,送上海英骏生物技术有限公司进行测序。所测得序列在NCBI上进行比对,并用MEGA软件采用邻接法(neighbor-joining method)构建系统发育树。
1.7 统计分析数据分析采用SPSS17.0统计分析软件,组间差异采用单因素方差分析(One way ANOVA)。数据以平均值±SE表示,P≤0.05表示存在显著性差异。
2 结果与分析 2.1 根际及非根际土壤真菌群落相似性分析如图 1所示,DGGE电泳图谱中可见数量众多强度不一的条带。其中,根际土壤样品电泳图谱中只有几条优势条带,更多的是强度较弱的条带。与根际土壤样品的DGGE图谱相比,非根际土壤样品DGGE电泳图谱中优势条带更少,仅有条带1和8两条优势条带。非根际土壤样品DGGE图谱的复杂度也较根际土壤样品小。条带1和8在根际和非根际土壤中均能检测到,且条带强度在各处理组中变化不大,表明它们所对应的真菌受环境影响较小。与之相比条带4和6在根际土壤DGGE图谱中强度较高,而在非根际土壤中变弱,说明它们所对应的真菌受植物根系影响较大。此外,还有一些条带如3和5仅存在于根际土壤DGGE图谱中。
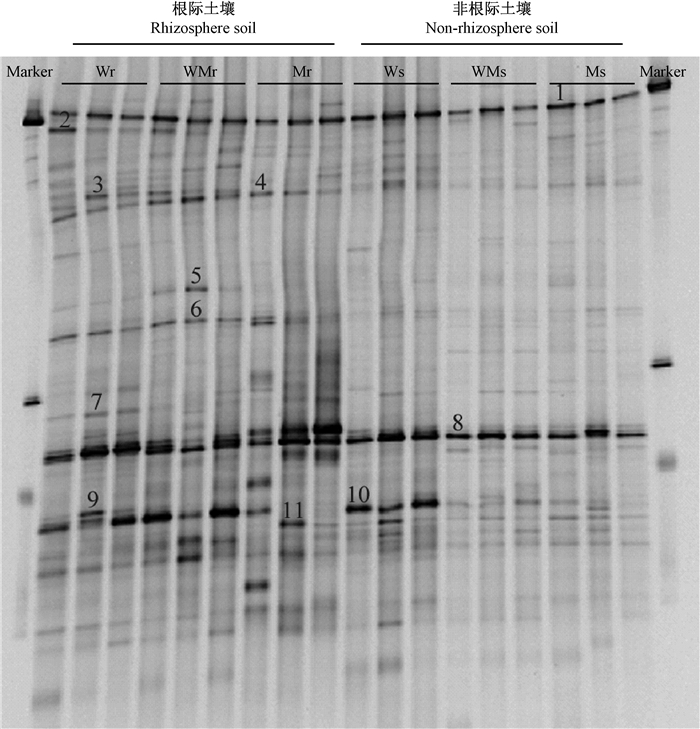 |
图 1 苦楝-小麦农林复合生态系统根际和非根际土壤真菌DGGE指纹图谱 Fig. 1 DGGE profiles of rhizosphere and bulk soil in M. azedarach-T.aestivum agroforestry ecosystem Wr:纯小麦根际土壤 Rhizosphere soil of wheat;WMr:苦楝-小麦间作模式中小麦根际土壤 Rhizosphere soil of wheat in M.azedarach-T.aestivum agroforestry ecosystem; ;Mr:苦楝根际土壤 Rhizosphere soil of M.azedarach; ;Ws:纯小麦非根际土壤 Non-rhizosphere soil of wheat; ;WMs:苦楝+小麦间作模式中小麦非根际土壤 Non-rhizosphere soil of T.aestivum in M.azedarac-T.aestivum agmforestry ecosystem; ;Ms:苦楝非根际土壤Non-rhizosphere soil of M.azedarach. |
进一步利用UPGMA进行聚类分析发现,所有样品明显聚为3簇(图 2)。第1组包括小麦根际土壤、小麦非根际土壤以及苦楝-小麦农林复合系统中的小麦根际土壤样品,相似度为68%,表明在此农林复合系统中小麦根际真菌群落受伴生树种苦楝的影响较小。第2组的相似度为65%,包括苦楝-小麦农林复合系统中的小麦非根际土壤样品和苦楝非根际土壤样品。第2组为苦楝根际土壤样品,其与第1,2二组的相似度为51%。说明苦楝根际真菌组成与其他处理相差较大。此外,同一处理的3个重复间相似性较大,除苦楝根际土壤样品外均在70%以上。
 |
图 2 苦楝-小麦农林复合生态系统真菌群落结构相似性的聚类分析 Fig. 2 Cluster analysis of fungal community structure similarity in M. azedarach-T.aestivum agroforestry ecosystem |
本研究中根际及非根际土壤真菌丰富度及多样性采用 S,H和E 3个指标进行综合评价,结果如图 3所示。由图 3A可知,就根际真菌丰富度而言,苦楝-小麦农林复合系统中的小麦根际真菌丰富度最高为23.33,而非根际土壤中小麦非根际土壤真菌丰富度最高为20.67,苦楝非根际土壤真菌丰富度最低为15.33,3组处理根际真菌之间以及非根际真菌间丰富度差异均不明显。进一步对同一处理组根际真菌与非根际真菌比较发现,小麦根际与非根际真菌丰富度无明显差异,而小麦-苦楝农林复合系统中的小麦及苦楝根际真菌丰富度显著高于非根际真菌(图 3D)。根际真菌的 H值存在显著差异, H值最高的是小麦-苦楝农林复合系统中的小麦根际真菌(3.04),最低的是苦楝非根际土壤真菌(2.48);非根际真菌的 H值差异不显著。与丰富度结果相似,小麦-苦楝农林复合系统中的小麦及苦楝根际真菌与非根际真菌的 H值均存在显著差异。根际真菌各组间以及非根际真菌各组间 E值差异不大,E值最高的是苦楝根际土壤真菌(0.97),最低的是苦楝非根际土壤真菌(0.92)。而小麦-苦楝农林复合系统中的小麦根际真菌 E值明显高于非根际真菌,同样苦楝根际真菌与非根际真菌 E值也存在明显差异。以上结果表明,小麦-苦楝农林复合生态系统中根际真菌多样性显著高于非根际真菌,这可能是系统中苦楝和小麦联合作用的结果。
 |
图 3 根际与非根际土壤真菌多样性分析 Fig. 3 Diversity analysis of fungi in the rhizosphere and non-rhizosphere soil |
引入主成分分析方法从而对复杂的DGGE图谱进行分析。如图 4所示,每个图标代表一个样品的菌群结构,图标之间的距离代表2个菌群间的差异,距离越大差异越大。PCA图在X轴解释了43.7%的变异度,在Y轴解释了17.9%的变异度。主成分分析进一步验证了聚类分析结果。由图 4可以看出,所有样品明显分为3个聚类群,其中苦楝非根际土壤真菌单独聚为一群。主成分分析结果进一步验证了聚类分析所得结果,即苦楝对小麦根际真菌群落的影响不大。
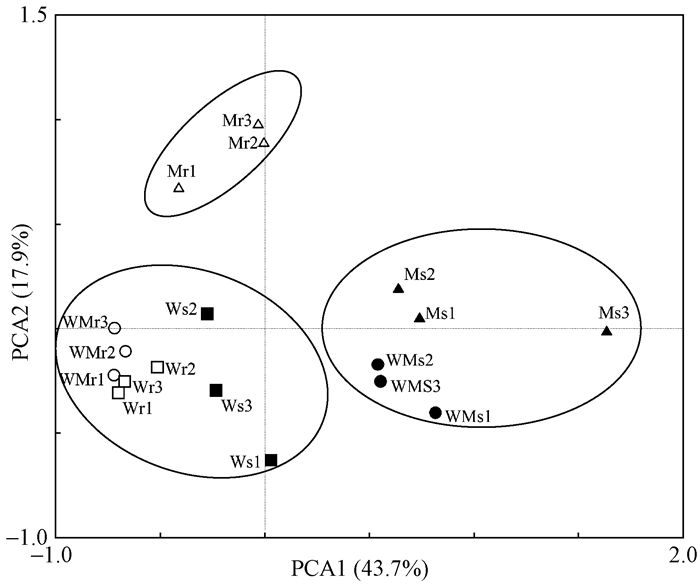 |
图 4 DGGE指纹图谱的主成分分析 Fig. 4 Principal component analysis of DGGE profiles |
对DGGE凝胶上的11条优势条带(图 1)进行割胶测序,并使用NCBI的BLAST工具与数据库内基因进行比对。如表 1和图 5所示,11条条带中有3条为不能培养的真菌,其余8条条带对应的同源性最高的序列分别属于球囊菌门(Glomeromycota)、担子菌门(Basidiomycota)、子囊菌门(Ascomycota)和半知菌亚门(Deuteromycota)。
|
|
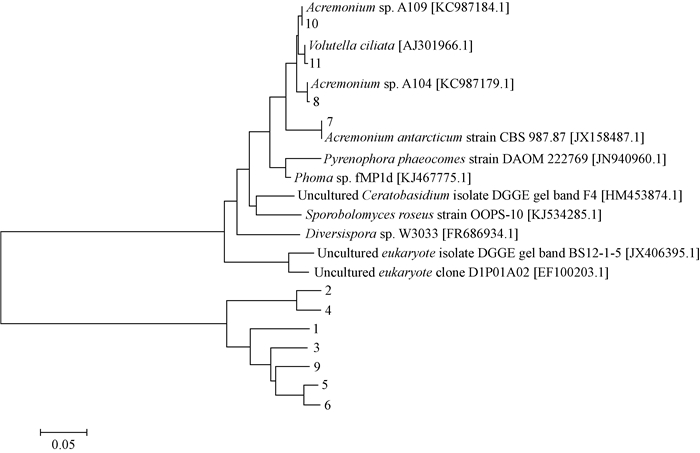 |
图 5 DGGE条带的系统发育树 Fig. 5 Phylogenetic tree of sequences derived from DGGE bands 方括号内为序列的GenBank登录号 GenBank numbers for the sequences were listed in square brackets. |
农林复合经营是一种环境友好的可持续土地利用方式,可以改变土壤的物理性状并能改善土壤质量(Rivest et al., 2013; Ketema et al., 2014)。土壤状况的改变进而会影响定居于此的各类微生物(Vallejo et al., 2012)。尽管已有一些研究涉及到土壤微生物对这种土地利用方式的反应,但这些研究主要集中在农林复合经营对微生物生物量的影响(Vallejo et al., 2012; Tangjang et al., 2009)。目前,尚未见将PCR-DGGE技术应用到农林复合生态系统土壤微生物群落组成与结构的相关报道。本研究中,笔者采用18S rDNA PCR-DGGE技术分析了苦楝-小麦农林复合生态系统根际及非根际土壤真菌群落结构与真菌组成。结果表明,在这一复合生态系统中根际与非根际土壤真菌群落结构间存在差异,特别是苦楝根际真菌群落结构与非根际土壤差异显著。此外,研究还发现小麦根际真菌与小麦-苦楝农林复合系统中的小麦根际真菌群落结构具有较高的相似性,表明苦楝对小麦根际真菌群落结构未造成较大影响。
本研究发现一些根际真菌的相对丰富度有所增加,具体表现为一些在非根际土壤样品DGGE谱图中较弱的条带在根际土壤样品DGGE谱图中强度明显增加,如条带4和6。笔者的研究结果与根际细菌群落的变化一致(Smalla et al., 2011)。这种根际效应在玉米 (Zea mays)根际真菌群落的变化过程中也已发现(Gomes et al., 2003)。究其原因,这种根际真菌的富集效应可能归因于植物根系分泌物。根系分泌物富含矿质离子、酶以及各种含碳化合物,这些物质可能在根际微生物种群富集过程中起重要作用(Berg et al., 2009; Uren,2000)。也有研究认为,这种根际效应是由于植物从土壤中选择特定的微生物到根际造成的(Singh et al., 2007)。还有学者推测,某些微生物种群在根际的富集不是由于植物的主动选择作用,而是因为这些微生物为避免被掠食而进入根际(Turner et al., 2013)。此外,笔者的研究结果显示,非根际土壤样品DGGE谱图中的优势条带明显少于根际土壤。大多数土壤中的微生物种群缺乏碳源供给(Garbeva et al., 2011),而植物将40%左右的光合产物分泌到根际(Bais et al., 2006),因此根际的微生物群落密度远远高于周围的非根际土壤。
植物根系会以一个物种特异的方式影响根际微生物群落(Berg et al., 2009),即不同植物会对其根际微生物产生不同影响。本研究结果表明苦楝根际真菌群落结构与小麦根际存在较大差异(相似度51%)。植物可以通过分泌一些物质选择性刺激或抑制某些微生物种群从而影响根际微生物群落结构(Doornbos et al., 2012)。此外,植物还可以通过分泌次生代谢产物抑制根际某些微生物的生长,如禾本科(Gramineae)作物的根部可以分泌苯并噁唑嗪酮类化合物抑制根际某些微生物(Berendsen et al., 2012)。因此,本研究中苦楝与小麦根际真菌群落结构的差异可能是由于两者根系分泌物的成分不同造成的。
PCR-DGGE分析技术已被广泛应用于环境微生物群落研究。该技术克服了传统微生物培养方法费时费力及对群落多样性的低估等缺陷。此外,DGGE技术可以在同一块凝胶上同时对多个样品进行分离和比较,分离的条带还可用来克隆测序(O′Callaghan et al., 2006)。尽管有诸多优点,DGGE技术也存在一定缺陷。DGGE分析技术的第一步是DNA的提取,此过程中如果微生物细胞裂解不充分会影响DNA产量,土壤中某些丰富度很低的种群可能会因此而被低估。PCR扩增过程中引物的选择会对DGGE结果有一定影响,而且不同批次凝胶电泳结果会有差异,从而造成试验重复稳定性较差。最近有研究者将焦磷酸测序技术引入森林生态系统土壤微生物群落研究(Hartmann et al., 2012),与DGGE技术相比焦磷酸测序可以对核酸序列进行快速准确的测定并且具有较高的覆盖度(Vaz-Moreira et al., 2011; Leite et al., 2012)。今后在农林复合生态系统菌群结构分析中可以将DGGE与焦磷酸测序技术相结合以获得更为准确的试验结果。本研究中采用了物种丰富度、香农多样性指数和均匀度指数3个指标对土壤真菌群落的遗传多样性进行综合评价,结果发现小麦-苦楝农林复合系统中的小麦根际真菌丰富度、多样性指数和均匀度均比单纯小麦根际真菌高,表明此复合系统中伴生的苦楝增加了小麦根际真菌的多样性。这一研究结果可为农林复合经营生产实践提供一些新的参考。
| [1] |
Bais H P, Weir T L, Perry L G, et al. 2006. The role of root exudates in rhizosphere interactions with plants and other organisms. Annual Review of Plant Biology, 57: 233-266.( 1) 1)
|
| [2] |
Berendsen R L, Pieterse C M J, Bakker P A H M. 2012. The rhizosphere microbiome and plant health. Trends in Plant Sciences, 17 (8): 478-486.( 2) 2)
|
| [3] |
Berg G, Smalla K. 2009. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiology Ecology, 68(1):1-13.( 3) 3)
|
| [4] |
Broeckling C D, Broz A K, Bergelson J, et al. 2008. Root exudates regulate soil fungal community composition and diversity. Applied and Environmental Microbiology, 74 (3):738-744.( 1) 1)
|
| [5] |
Doornbos R F, van Loon L C, Bakker P A H M. 2012. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agronomy for Sustainable Development, 32 (1): 227-243.( 1) 1)
|
| [6] |
Fanish S A, Priya R S. 2013. Review on benefits of agroforestry system. International Journal of Education and Research, 1(1):1-12.( 1) 1)
|
| [7] |
Finlay R D. 2008. Ecological aspects of mycorrhizal symbiosis: with special emphasis on the functional diversity of interactions involving the extraradical mycelium. Journal of Experimental Botany, 59 (5): 1115-1126.( 1) 1)
|
| [8] |
Garbeva P, Hol W H G, Termorshuizen A J, et al. 2011. Fungistasis and general soil biostasis-a new synthesis. Soil Biology and Biochemistry, 43 (3): 469-477.( 1) 1)
|
| [9] |
Gomes N C M, Fagbola O, Costa R, et al. 2003. Dynamics of fungal communities in bulk and maize rhizosphere soil in the tropics. Applied and Environmental Microbiology, 69 (7): 3758-3766.( 1) 1)
|
| [10] |
Haichar F Z, Marol C, Berge O, et al. 2008. Plant host habitat and root exudates shape soil bacterial community structure. The ISME Journal, 2(12): 1221-1230.( 1) 1)
|
| [11] |
Hartmann M, Howes C G, VanInsberghe D, et al. 2012. Significant and persistent impact of timber harvesting on soil microbial communities in Northern coniferous forests. The ISME Journal, 6(12): 2199-2218.( 1) 1)
|
| [12] |
Hinsinger P, Bengough A G, Vetterlein D, et al. 2009. Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant and Soil, 321 (1):117-152.( 1) 1)
|
| [13] |
Ketema H, Yimer F. 2014. Soil property variation under agroforestry based conservation tillage and maize based conventional tillage in Southern Ethiopia. Soil and Tillage Research, 141: 25-31.( 2) 2)
|
| [14] |
Khan A L, Hamayun M, Kang S M, et al. 2012. Endophytic fungal association via gibberellins and indole acetic acid can improve plant growth under abiotic stress: an example of Paecilomyces formosus LHL10. BMC Microbiology, 12: 3.( 1) 1)
|
| [15] |
Leite A M O, Mayo B, Rachid C T C C, et al. 2012. Assessment of the microbial diversity of Brazilian kefir grains by PCR-DGGE and pyrosequencing analysis. Food Microbiology, 31 (2): 215-221.( 1) 1)
|
| [16] |
Manici L M, Caputo F. 2010. Soil fungal communities as indicators for replanting new peach orchards in intensively cultivated areas. Europe Journal of Agronomy, 33 (3):188-196.( 1) 1)
|
| [17] |
Mendes R, Garbeva P, Raaijmakers J M. 2013. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganism. FEMS Microbiology Reviews, 37: 634-663.( 2) 2)
|
| [18] |
O'Callaghan M, Lorenz N, Gerard E M. 2006. Characterization of phylloplane and rhizosphere microbial populations using PCR and denaturing gradient gel electrophoresis (DGGE)//Cooper J E, Rao J R. Molecular approaches to soil, rhizosphere and plant microorganism analysis. CABI International, Oxfordshire, UK, 99-115.( 1) 1)
|
| [19] |
Ogino A, Koshikawa H, Nakahara T, et al. 2001. Succession of microbial communities during a biostimulation process as evaluated by DGGE and clone library analyses. Journal of Applied Microbiology, 91 (4): 625-635.( 1) 1)
|
| [20] |
Rivest D, Lorente M, Olivier A, et al. 2013. Soil biochemical properties and microbial resilience in agroforestry systems: Effects on wheat growth under controlled drought and flooding conditions. Science of The Total Environment, 463,464: 51-60.( 2) 2)
|
| [21] |
Saito A, Ikeda S, Ezura H, et al. 2007. Microbial community analysis of the phytosphere using culture-independent methodologies. Microbes and Environments, 22 (2): 93-105.( 1) 1)
|
| [22] |
Singh B K, Munro S, Potts J M, et al. 2007. Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Applied Soil Ecology, 36 (2/3): 147-155.( 1) 1)
|
| [23] |
Smalla K, Wieland G, Buchner A, et al. 2011. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Applied and Environmental Microbiology, 67 (10): 4742-4751.( 1) 1)
|
| [24] |
Tangjang S, Arunachalam K, Arunachalam A, et al. 2009. Microbial population dynamics of soil under traditional agroforestry systems in northeast India. Research Journal of Soil Biology, 1 (1): 1-7.( 1) 1)
|
| [25] |
Teixeira L C R S, Peixoto R S, Cury J C, et al. 2010. Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. The ISME Journal, 4(8): 989-1001.( 1) 1)
|
| [26] |
Turner T R, Ramakrishnan K, Walshaw J, et al. 2013. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. The ISME Journal, 7(12): 2248-2258.( 1) 1)
|
| [27] | Urbanová M, Šnajdr J, Baldrian P. 2015. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biology and Biochemistry, 84: 53-64. |
| [28] |
Uren N C. 2000. Types, amounts and possible functions of compounds released into the rhizosphere by soil grown plants//Pinton R, Varani Z, Nanniperi P. The rhizosphere: biochemistry, and organic substances at the soil interface. Marcel Dekker Inc., New York, 19-40.( 1) 1)
|
| [29] |
Vainio E J, Hantula J. 2000. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycological Research, 104 (8): 927-936.( 1) 1)
|
| [30] |
Vallejo V E, Arbeli Z, Terán W, et al. 2012. Effect of land management and Prosopis juliflora (Sw.) DC trees on soil microbial community and enzymatic activities in intensive silvopastoral systems of Colombia. Agriculture, Ecosystems & Environment, 150: 139-148.( 2) 2)
|
| [31] |
Vaz-Moreira I, Egas C, Nunes O C, et al. 2011. Culture-dependent and culture-independent diversity surveys target different bacteria: a case study in a freshwater sample. Antonie van Leeuwenhoek, 100 (2): 245-257.( 1) 1)
|
| [32] |
Yu Z T, Morrison M. 2004. Comparisons of different hypervariable regions of rrs genes for use in fingerprinting of microbial communities by PCR-denaturing gradient gel electrophoresis. Applied and Environmental Microbiology, 70 (8): 4800-4806.( 1) 1)
|
 2015, Vol. 51
2015, Vol. 51

