文章信息
- 李科友, 朱海兰
- Li Keyou, Zhu Hailan
- 植物非生物逆境胁迫DREB/CBF转录因子的研究进展
- Research Progress of DREB/CBF Transcription Factor in Response to Abiotic-Stresses in Plants
- 林业科学, 2011, 47(1): 124-134.
- Scientia Silvae Sinicae, 2011, 47(1): 124-134.
-
文章历史
- 收稿日期:2010-08-02
- 修回日期:2010-10-06
-
作者相关文章
2. 西北农林科技大学林学院 杨凌 712100
2. College of Forestry, Northwest A & F University Yangling 712100
干旱、盐碱、低温等非生物逆境是影响植物生长发育的主要因素,植物受到逆境胁迫时会产生形态、生理、基因表达等适应性调节反应以降低或消除危害。转录因子(反式作用因子)基因是植物中最重要的一类调节基因,其在植物体内构成复杂的调节网络, 在时间和空间上协同控制基因的表达。转录因子是能够与真核基因启动子区域中顺式作用元件发生特异性作用的DNA结合蛋白,通过它们之间以及与其他相关蛋白之间的相互作用,激活或抑制基因转录。DREB/CBF (Dehydration Responsive Element Binding Protein/C-repeat Binding Factor)转录因子,即干旱应答元件结合蛋白质/C-重复序列结合子,能特异结合DRE/CRT(Dehydration Responsive Element /C-repeat)顺式作用元件。DRE/CRT顺式作用元件普遍存在于干旱、高盐或低温等逆境应答基因的启动子中,核心序列为CCGAC。DREB/CBF转录因子由逆境胁迫诱导产生后,可激活其他一系列依赖DRE/CRT顺式作用元件的抗逆功能基因的表达,从而增强植物对干旱、低温及高盐等逆境的抗性(Agarwal et al., 2006)。因此,DREB/CBF转录因子在植物抗逆中的作用越来越广泛地受到重视,目前,已成为植物抗逆分子生物学研究的热点之一。本文就近十几年特别是近5年来国内外DREB/CBF转录因子的研究进展, DREB/CBF转录因子在植物抗逆中的作用及研究作一综述,旨在为植物抗逆育种提供理论依据。
1 DREB/CBF转录因子的结构特点迄今报道的DREB/CBF转录因子基因内均无内含子,从蛋白质结构分析,DREB/CBF转录因子含有1个保守的AP2/EREBP结构域,是转录因子AP2/EREBP家族中的1个亚家族, 主要调节植物对低温、干旱和高盐等逆境的分子应答。DREB/CBF亚族又分为6个亚组(A-1—A-6),氨基酸序列对比发现, A-1类转录因子上游含有1个保守的核定位信号区(NLS): PKRPAGRTKFRETRHP,下游含有DSAW的保守序列和羧基末端的LWSY保守序列, 这些序列被称为A-1类转录因子的特征序列(Jaglo et al., 2001)。而在与干旱、高盐诱导相关的A-2类转录因子中不存在(Yamaguchi et al., 2006)。A-3—A-6 4个亚组占整个拟南芥(Arabidopsis thaliana) DREB/CBF亚族基因的75%,但目前对它们的功能研究很少。最近,对这4个DREB/CBF亚组的基因研究相继有一些报道, 如拟南芥TINY2(A-4) (Wei et al., 2005), 大豆(Glycine max)Gm-DREB2(A-5) (Chen et al., 2007), 陆地棉(Gossypium hirsutum)GhDBP3(A-4), (A-5), GhDBP2(A-6) (Huang et al., 2006a)和玉米(Zea mays)ZmDBF1(A-6) (Kizis et al., 2002)。
DREB/CBF转录因子的二级结构具有典型的结构特征:在C-末端富含酸性氨基酸,只有少量碱性氨基酸,功能是作为转录激活区。例如小麦(Triticum aestivum)的TaDREB1 C-末端的50个氨基酸中酸性氨基酸占25%,碱性氨基酸仅为4%;榆钱菠菜(Atriplex hortensis)的AhDREB1 C-末端的80个氨基酸中酸性氨基酸占23%,碱性氨基酸仅为3%(Shen et al., 2003a;2003b)。N-末端富含碱性氨基酸,是核定位信号区;中间由58个左右氨基酸残基组成的AP2/EREBP结构域,分为2个保守元件YRG和RAYD。YRG含有19~22个氨基酸残基, 大多数为碱性氨基酸残基, 有利于与DNA的结合。RAYD由42或43个氨基酸残基组成, 含有由18个氨基酸组成的α-双亲螺旋的核心区域;AP2/EREBP结构域可形成3个β-折叠和1个α-螺旋结构,其中位于第2个β-折叠中的第14位的缬氨酸(V)和第19位的谷氨酸(E),特别是第14位的缬氨酸(V),对决定DREB/CBF转录因子与DRE/CRT顺式作用元件的特异性结合起关键作用(Liu et al., 1998;Sakuma et al., 2002)。已报道的DREB/CBF转录因子的AP2/EREBP结构域中有7个关键残基参与了DREB/CBF蛋白与DRE/CRT顺式作用元件的直接作用,它们是4个精氨酸(R)残基、2个色氨酸(W)残基和1个缬氨酸(V)残基(陈金焕等,2007)。Gao等(2002)通过突变的方法发现缬氨酸(V)的突变导致DREB/CBF蛋白结合DRE/CRT顺式作用元件的活性基本丧失,从而说明缬氨酸(V)是DREB/CBF家族的特征残基。
2 DREB/CBF转录因子基因的克隆自Stockinger等(1997)首次从拟南芥cDNA文库中克隆到AtCBF1以来,DREB/CBF转录因子基因的克隆得到较大的发展,已陆续从各种植物中克隆出DREB/CBF基因(表 1)。
|
|
Stockinger等(1997)利用酵母一元杂交方法首次从拟南芥cDNA文库中克隆到DRE/CRT结合蛋白质基因,并命名为CBF1 (CRT-binding factor 1)。Liu等(1998)利用rd29A基因启动子的DRE/CRT顺式作用元件和酵母一元杂交方法,从拟南芥中克隆了2类共5个与DRE元件特异结合,在低温、干旱或高盐胁迫下调控基因表达的DREB转录因子,分别命名为DREB1A (CBF3),DREB1B(CBF1),DREB1C(CBF2)和DREB2A,DREB2B。自那时以来,DREB/CBF转录因子一直是植物分子生物学研究的热点。
植物的干旱、高盐及低温胁迫应答途径涉及到依赖于ABA和不依赖ABA的信号传导途径。目前,从植物中分离得到的DREB/CBF类转录因子都能通过与DRE/CRT元件的相互作用,完成对干旱、高盐或低温胁迫应答基因的激活作用。其中A-1和A-2组的主要成员DREB1A/B/C和DREB2转录因子分别参与到不依赖于ABA的低温和脱水(干旱或高盐)胁迫应答途径中,其他受逆境诱导的DREB/CBF转录因子如DREB1D/CBF4, GmDREBa,GmDREB2,ZmDBP2,ZmDBF1, CkDBF,MrDREBA6,TaDREBA6,PpDBF1,DvDREB,AvDREB2,GhDBP2和GhDBP3等参与到依赖ABA的脱水应答途径中。这些结果表明不同植物中类似的DREB/CBF基因, 其参与的细胞内信号转导的途径可能不同,植物在逆境胁迫应答反应中存在着复杂的信号传递途径,并且各种胁迫信号传递途径间通过某些共同的组分联系在一起,构成一个复杂的信号传递网络(Haake et al., 2002)。大部分DREB/CBF基因的表达受干旱、盐碱、低温或冻害等逆境胁迫的诱导,但不同形式DREB/CBF在同一植物和不同植物中诱导表达的条件和所需时间不同。
在正常生长条件下,在拟南芥中几乎检测不到AtDREB1基因转录产物的存在;当受到低温协迫时,AtDREB1A, AtDREB1B和AtDREB1C基因能在15 min内表达,并在2 h内达到最高,随后开始下降,但在24 h内基因表达仍然高于对照;而对于AtDREB2基因来说,干旱和高盐处理时,10 h表达水平达到最高(Liu et al., 1998)。AtCBF4基因被干旱和ABA处理快速强烈诱导,而不受低温诱导(Haake et al., 2002)。水稻低温处理后40 min内OsDREB1A和OsDREB2A基因被快速诱导,但不受外源ABA的诱导。OsDREB1A盐处理5 h后被诱导, OsDREB1C为组成型表达,OsDREB1D在胁迫和正常条件下检测不到,OsDREB2A干旱或高盐250 mmol·L-1 NaCl处理24 h被诱导,对低温和ABA处理反应不敏感(Dubouzet et al., 2003)。辣椒(Capsium annuum)中CaDREBLP1被干旱、高盐快速诱导,机械损伤也可诱导,但不被低温诱导,这种表达方式与AtDREB2A表达方式十分相似,但还属于DREB1类基因,因为其结构特征与其他DREB1基因类似,这说明CaDREBLP1属于一种新型的DREB基因(Hong et al., 2005)。
DREB/CBF转录因子家族可以特异性地识别并结合DRE/CRT顺式作用元件,调控RD29A,RD17,ERD10,KIN1,COR6.6,KIN2和COR15A等多种下游基因(启动子区域含有DRE/CRT顺式作用元件)的表达,其自身的表达也受上游基因作用、“同伴”作用以及其下游基因的反馈作用。研究表明,DREB/CBF的上游基因ICE1(inducer of CBF expression 1),LOS1(low expression of osmotically respessive genes 1),LOS4(low expression of osmotically respessive genes 4),FRY2(fiery2),HOS1(high expression of osmotically respessive genes 1)等对DREB/CBF的表达都起着重要的调控作用(Chinnusamy et al., 2003;Lee et al., 2001)。Novillo等(2004)通过基因芯片技术发现, 在低温情况下,AtDREB1A/AtCBF3和AtDREB1B/AtCBF1较AtDREB1C/AtCBF2提前表达。此外, 在拟南芥cbf2突变株中, AtDREB1A/AtCBF3和AtDREB1B/AtCBF1的表达量都有所增加, 其抗旱耐盐抗冻的能力也较野生型强。这些研究说明AtDREB1C/AtCBF2对AtDREB1A/AtCBF3和AtDREB1B/AtCBF1起到了反向调节的作用(Novillo et al., 2004)。Gao等(2002)发现在欧洲油菜(Brassica napus)los1突变株中, 由低温诱导的含有DRE/CRT元件的基因合成受阻时CBF的表达量却增高, 推测CBF/DREB1可能被自己的基因产物和下游基因产物所反馈抑制。
有关特定组织中DREB/CBF表达的研究还不多。在正常生长条件下,AtDREB2A和AhDREB1在拟南芥和山波菜根、茎和叶中均得到表达,在盐胁迫下,AhDREB1在根中高度表达,但在茎叶中表达量差异不大(Shen et al., 2003b)。在大豆幼叶中GmDREBa被低温、干旱、高盐快速诱导,而GmDREBb的表达则不明显,但干旱、盐和ABA处理后GmDREBc在根中表达程度很高(Li et al., 2005)。菊花(Dendranthema morifolium) DmDREBa和DmDREBb在茎和叶中的表达量比在根和花中的表达量高(Liu et al., 2008)。番薯(Ipomoea batatas)SwDREB1在茎和块状根中高水平表达(Kim et al., 2008)。西府海棠(Malus micromalus) MrDREB6在叶中的表达量远高于其他组织,其次在种子中,在茎中的表达量最少(付晓燕等,2009)。这种组织特异性表达可能与植物的不同抗性机制有关,受启动子特异元件调控。
植物细胞从感应干旱、高盐或低温胁迫开始,通过各级信号传递DREB/CBF转录因子与DRE/CRT顺式作用元件相互作用的转录调控,目的基因的表达、基因产物的积累引起的各种生理生化调节,最终使植物抗逆性获得增强,这一系列过程可用图 1来概括(刘强等,2000)。
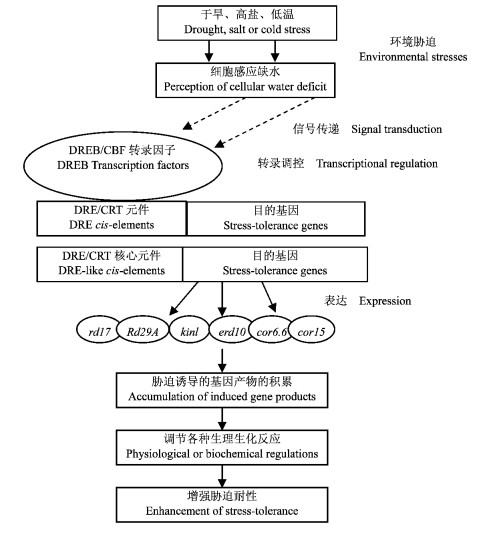
|
图 1 逆境胁迫下DREB/CBF转录因子的感应和表达调控过程 Figure 1 A process of induction, expression and regulation of DREB/CBF transcription factor in environmental stresses |
由于植物对低温、干旱和高盐等逆境胁迫耐性受多个基因的控制,因此其强弱往往不取决于单个因子,而是受许多因子影响。DREB/CBF转录因子能够识别DRE/CRT元件,与基因启动子区域的DRE/CRT顺式作用元件特异性结合,在信号传导及诱导下游许多抗逆相关的功能基因表达中起到关键作用。因此,利用转录因子增强植物抗逆性成为改良植物抗逆性的重要途径。DREB/CBF转录因子自首次报道以来,一直受到广泛关注,研究者们通过转基因手段鉴定DREB/CBF转录因子的功能,同时也得到了许多抗性增强的转基因植株(表 2)。
|
|
DREB/CBF转录因子基因在转基因植物中表达较为复杂。目前,在植物抗逆基因工程中主要利用组成性启动子和特异诱导性启动子构建DREB/CBF转基因抗逆品系。
利用组成型CaMV35S启动子在拟南芥中过量表达的AtDREB1A, OsDREB1A或GmDREB2 (Kasuga et al., 1999;Dubouzet et al., 2003;Chen et al., 2007), 在水稻中过量表达的AtDREB1A(Oh et al., 2005)和在烟草中过量表达的AhDREB1, GhDREB1(Shen et al., 2003a;Shan et al., 2007)均能增强植物抗逆性,但在一些情况下过量表达的DREB/CBF导致植株生长矮化(Liu et al., 1998;Kasuga et al., 1999;Dubouzet et al., 2003)。这可能是由于组成型CaMV35S启动子的控制,外源基因在转基因植物所有发育阶段都会持续表达,造成植物体内资源的非必要浪费,此外大量外源蛋白的积累也会干扰植物正常的代谢调控;使用特异诱导性rd29A启动子可以消除矮化现象(Kasuga et al., 1999;Pellegrineschi et al., 2004;Sakuma et al., 2006)说明诱导型启动子rd29A能更有效地提高植物对低温、干旱和高盐的耐受性(Kasuga et al., 1999;Agarwal et al., 2006),因为诱导型启动子是在遇到胁迫信号时才启动基因的表达,不会影响植物体在正常情况下的生长发育。
Hsieh等(2002a)将与CaMV35S启动子连接的AtCBF1基因转入番茄(Lycopersicon esculentum),发现转基因番茄体内有类似拟南芥中的DREB信号转导过程,同时发现自由基减少,脯氨酸含量升高,植株的抗旱性和耐低温性都明显增强,但是也出现生长矮化现象;用GA3处理后,植株恢复正常生长,且耐寒能力仍明显高于野生型植株。但是CaMV35S启动子驱动AtDREB1A/AtCBF3过量表达的转基因番茄和拟南芥,用GA3处理后,矮化现象并不发生变化(Hsieh et al., 2002a;2002b)。因此,转基因植株产生的矮化现象可能并非GA生物合成干扰,而是其他机制所致。
将AtDREB2A基因转入到拟南芥中,过度表达AtDREB2A的转基因拟南芥没有增强对逆境的抗性,由此推测AtDREB2A基因产物激活下游靶基因可能需要磷酸化或去磷酸化修饰(Liu et al., 1998;Dubouzet et al., 2003)。Sakuma等(2006)研究发现在拟南芥AtDREB2A基因中第136~165位氨基酸残基抑制了DREB2A蛋白质的活性,将该区域删除后,过度表达DREB2A的转基因拟南芥提高了对干旱的抗性。Agarwal等(2007)研究发现狼尾草(Pennisetum glaucum) PgDREB2A是一个磷酸化蛋白, 并且磷酸化作用负调控蛋白对DNA的结合活性, 磷酸化后的蛋白不能结合DRE元件, 当用磷酸酶处理后, PgDREB2A结合DRE元件的能力得到了恢复。但是,Dubouzet等(2003)在另一个试验中,分别将拟南芥和水稻的OsDREB2A基因转入到水稻原生质体中却激活了GUS基因的表达。所以,DREB2A基因产物激活下游靶基因需要磷酸化或去磷酸化,这一结论并不是转基因植株胁迫抗性没有提高的真正原因,探明DREB2A基因如何提高植物的抗性还需要做进一步的研究。
Chen等(2007)从大豆(Glycine max)中克隆得到受干旱、高盐、低温以及ABA诱导的转录因子GmDREB2基因,将其分别置于特异启动子rd29A和CaMV35S启动子下转入拟南芥后,2种转基因植株的生长情况和野生型植株相比均未表现出异常,逆境处理试验表明转基因植株的抗性明显提高。目前,已从大豆中分离出6个DREB同源基因GmDREBa,GmDREBb,GmDREBc,GmDREB1,GmDREB2和GmDREB3 (Li et al., 2005; Chen et al., 2007;2009),其中GmDREB1, GmDREB2和GmDREB3同属于DREB家族的A-5亚家族,过量表达的GmDREB1,GmDREB2和GmDREB3可以增强转基因拟南芥植株的抗旱、抗低温和抗高盐性(Chen et al., 2007;2009;Jin et al., 2010),这表明DREB家族的亚家族A-5是提高植物抗逆性的重要基因资源。
总之,一系列的转基因研究结果表明,DREB/CBF转录因子家族不论是在双子叶植物、单子叶植物, 还是在草本植物及木本植物抗逆品种改良中均具有十分重要的应用价值。
5 DREB/CBF转录因子研究中的问题与展望综上所述,DREB/CBF在植物逆境信号转导途径中具有重要的位置,已对DREB/CBF的结构、类型、表达特性,在逆境胁迫下的作用及在培育转基因抗逆植物方面进行了大量的研究工作,这对于深刻理解植物的抗逆机制及利用该机制进行抗逆新品种的培育有重要意义,但依然存在许多问题有待进一步研究。
第一, DREB/CBF对于上游调控基因的研究还有待加强。Chinnvsamy等(2003)在拟南芥中发现了一个转录因子ICE1, 该基因编码1个MYC-type bHLH(basic helix-loop-helix)蛋白, 能够调节DREB1A/CBF3基因的表达,但是对其他DREB1/CBF基因没有明显的调节作用。冷信号激活ICE1表达的机制,ICE1在冷信号和DREB1/CBF之间的具体调节过程,细胞膜上特异冷信号受体的分离鉴定,理清包括Ca2+作为第2信使瞬时积累到ICE1的转录后修饰之间的上游事件等,还有待以后研究。
第二, 有关DREB/CBF类抗逆境转录因子的功能及转录调控的研究大部分来自拟南芥,而对其他植物的研究相对较少。Benedict等(2006)发现杨树和拟南芥中的调节子(regulon)相似, 但是对这些调节子的上游序列进行分析, 并没有发现DRE(CCGAC)的大量存在, 而ABRE却比较丰富, 这2个顺式元件在杨树调节子中的相互作用关系也有待继续研究。已完成的杨树基因组测序为木本植物DREB/CBF调控途径的研究提供了有力的条件,对DREB/CBF在木本植物中的调控网络进行解析,并将其应用于优质抗逆林木的筛选,将是今后研究的一个重要方向。
第三, 已对DREB/CBF类抗逆境转录因子在干旱、低温和高盐胁迫的应答进行了大量的研究,而对它们在其他胁迫如高温、高湿和抗氧化等胁迫方面的研究较少;DREB/CBF转录因子识别各种不同顺式作用元件并与之结合的机制,DREB/CBF转录因子影响转录起始速度的机制和如何接受外源信号以及如何控制靶基因的表达还不够清晰。
第四,转入DREB/CBF基因受体植物种类不够广泛,尚缺乏能够在田间应用的抗逆转基因植物。例如转CBF基因时,所研究的对象主要集中在拟南芥、欧洲油菜、番茄、大麦、小麦等植物上,对于一些亲缘关系较远的植物是否有效,还有待进一步研究;有效调控外源DREB/CBF基因表达,目前除了rd29A基因启动子控制DREB/CBF基因外,缺乏更多的探索。
DREB/CBF转录因子所介导的基因表达调控网络在植物抵御各种环境胁迫的应答中具有重要功能,在感知胁迫信号到胁迫应答基因表达的整个信号传导网络中,各种转录因子和顺式元件间的相互作用构成了基因表达调控的基础。植物对胁迫应答的反应是由多基因协同控制的。利用生物技术改良农林作物的抗逆性, 针对性强,效率高。在抗逆分子育种中, 与导入或改良个别功能基因来提高某种抗性的方法相比, 导入或改良一个转录因子是提高植物抗逆性更为有效的方法和途径。鉴于DREB/CBF转录因子在植物抗逆中的特殊地位,相信在不远的将来,随着对其在植物抗逆中分子机制的深入研究,DREB/CBF转录因子在培育抗逆植物新品种方面会取得新的重要进展,将有巨大的应用前景。
程汉, 安泽伟, 黄华孙. 2005. 巴西橡胶树CBF1基因的克隆及序列分析[J]. 热带作物学报, 26(3): 50-55. |
付晓燕, 彭日荷, 章镇, 等. 2009. 八棱海棠中转录因子基因MvDREBA6的克隆及表达分析[J]. 果树学报, 26(6): 761-768. |
高峰, 高强, 岳桂东, 等. 2005. 小盐芥(Thellungiella salsuginea) CBF1基因的克隆[J]. 山东大学学报, 40(5): 113-118. |
高世庆, 徐惠君, 程宪国, 等. 2005. 转大豆GmDREB基因增强小麦的耐旱及耐盐性[J]. 科学通报, 50(23): 22617-22627. |
洪波, 仝征, 马男, 等. 2006. AtDREB1A基因在菊花中的异源表达提高了植株对干旱和盐渍胁迫的耐性[J]. 中国科学, 36(3): 223-231. |
刘强, 赵南明, Yamaguch-ShinozakiK, 等. 2000. DREB转录因子在提高植物抗逆中的作用[J]. 科学通报, 45(1): 11-16. |
倪志勇, 徐兆师, 刘丽, 等. 2008. 小麦转录因子TaDREB6基因的克隆及鉴定[J]. 麦类作物学报, 28(3): 357-363. DOI:10.7606/j.issn.1009-1041.2008.03.086 |
秦红霞, 贾志平, 张海超, 等. 2005. 银新杨中与DRE元件结合的转录因子的克隆及鉴定分析[J]. 生物工程学报, 21(6): 906-910. |
吴关庭, 郎春秀, 胡张华, 等. 2006. 转CBF1基因增强水稻的耐逆性[J]. 核农学报, 20(3): 169-173. |
谢永丽, 王自章, 刘强, 等. 2005. 草坪草狗牙根中抗逆基因BeDREB的克隆及功能鉴定[J]. 中国生物化学与分子生物学报, 21(4): 521-527. |
阳文龙, 刘敬梅, 刘强, 等. 2006. 高羊茅DREB类转录因子基因的分离及鉴定分析[J]. 核农学报, 20(3): 187-192. |
杨春霞, 李火根, 程强, 等. 2009. 南林895杨抗旱耐盐基因DREB1C的转化[J]. 林业科学, 45(2): 17-21. DOI:10.11707/j.1001-7488.20090203 |
杨风萍, 梁荣奇, 张立全, 等. 2006. 抗逆调节转录因子CBF1基因提高多年生黑麦草的抗旱能力[J]. 华北农学报, 21(1): 14-18. |
张梅, 刘炜, 毕玉平, 等. 2009. 花生中DREB类转录因子PNDREB1的克隆及鉴定[J]. 作物学报, 35(11): 1973-1980. |
张倩, 马婧, 何婧, 等. 2009. 中国芦荟AlDREBA2基因的克隆及胁迫表达[J]. 园艺学报, 36(11): 1659-1666. DOI:10.3321/j.issn:0513-353X.2009.11.014 |
甄伟, 陈溪, 孙思洋, 等. 2000. 冷诱导基因的转录因子CBF1转化油菜和烟草及抗寒鉴定[J]. 自然科学进展, 10(12): 1104-1108. DOI:10.3321/j.issn:1002-008X.2000.12.009 |
周洲洲, 李永丽. 2010. 毛白杨转录因子PtCBF5的表达模式分析[J]. 林业科学, 46(4): 58-53. DOI:10.11707/j.1001-7488.20100409 |
Agarwal P K, Agarwal P, Reddy M K, et al. 2006. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants[J]. Plant Cell Rep, 25(12): 1263-1274. DOI:10.1007/s00299-006-0204-8 |
Agarwal P, Agarwal P K, Nair S, et al. 2007. Stress-inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA-binding activity[J]. Mol Genet Genomics, 277(2): 189-198. DOI:10.1007/s00438-006-0183-z |
Agarwal P, Agarwal P K, Joshi A J, et al. 2010. Overexpression of PgDREB2A transcription factor enhances abiotic stress tolerance and activates downstream stress-responsive genes[J]. Mol Biol Rep, 37(2): 1125-1135. DOI:10.1007/s11033-009-9885-8 |
Behnam B, Kikuchi A, Celebi-Toprak F, et al. 2006. The Arabidopsis DREB1A gene driven by the stress-inducible rd29A promoter increases salt-stress tolerance in tetrasomic tetraploid potato (Solanum tuberosum)in proportion to its copy number[J]. Plant Biotechnol, 23(2): 169-177. DOI:10.5511/plantbiotechnology.23.169 |
Behnam B, Kikuchi A, Celebi-Toprak F, et al. 2007. Arabidopsis rd29A::DREB1A expression of At DREB1A enhances freezing tolerance in transgenic potato[J]. Plant Cell Rep, 26(8): 1275-1282. DOI:10.1007/s00299-007-0360-5 |
Benedict C, Skinner J S, Meng R, et al. 2006. The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserve in Populus spp[J]. Plant Cell Environ, 29(7): 1259-1272. DOI:10.1111/pce.2006.29.issue-7 |
Bhatnagar-Mathur P, Devi M J, Reddy D S, et al. 2007. Stress-inducible expression of At DREB1A in transgenic peanut (Arachis hypogaea L.) increases transpiration efficiency under water-limiting conditions[J]. Plant Cell Rep, 26(12): 2071-2082. DOI:10.1007/s00299-007-0406-8 |
Cao Z F, Li J, Chen F, et al. 2002. Effect of two conserved amino acid residues on DREB1A function[J]. Biochemistry(Moscow), 66(6): 623-627. |
Celebi-Toprak F, Behnam B, Serrano G, et al. 2005. Tolerance to salt stress in transgenic tetrasomic tetraploid potato, Solanum tuberosum cv.Desiree appears to be induced by the DREB1A gene and rd29A promoter of Arabidopsis thaliana[J]. Breed Sci, 55(3): 311-319. DOI:10.1270/jsbbs.55.311 |
Chen J Q, Dong Y, Wang Y J, et al. 2003. An AP2/EREBP-type transcription-factor gene from rice is coldinducible and encodes a nuclear-localized protein[J]. Theor Appl Genet, 107(6): 972-979. DOI:10.1007/s00122-003-1346-5 |
Chen Jiren, Lü Jingjing, Liu Rong, et al. 2010. DREB1C from Medicago truncatula enhances freezing tolerance in transgenic M.truncatula and China Rose (Rosa chinensis Jacq.)[J]. Plant Growth Regul, 60(3): 199-211. DOI:10.1007/s10725-009-9434-4 |
Chen M, Wang Q Y, Cheng X G, et al. 2007. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants[J]. Biochemical and Biophysical Research Communications, 353(2): 299-305. DOI:10.1016/j.bbrc.2006.12.027 |
Chen M, Xu Z S, Xia L Q, et al. 2009. Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.)[J]. J Exp Bot, 60(1): 121-135. DOI:10.1093/jxb/ern269 |
Chinnusamy V, Ohta M, Kanrar S, et al. 2003. ICE1:a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis[J]. Genes Dev, 17(8): 1043-1054. DOI:10.1101/gad.1077503 |
Choi D W, Rodriguez E M, Close T J. 2002. Barley Cbf3 gene identification, expression pattern, and map location[J]. Plant Physiol, 129(4): 1781-1787. DOI:10.1104/pp.003046 |
Dubouzet J G, Sakuma Y, Ito Y, et al. 2003. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought, high-salt and cold-responsive gene expression[J]. Plant J, 33(4): 751-763. DOI:10.1046/j.1365-313X.2003.01661.x |
Egawa C, Kobayashi F, Ishibashi M, et al. 2006. Differential regulaion of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat[J]. Gene Genet Syst, 81(2): 77-91. DOI:10.1266/ggs.81.77 |
Gamboa M C, Rasmussen-Poblete S, Valenzuela P D, et al. 2007. Isolation and characterization of a cDNA encoding a CBF transcription factor from E.globules[J]. Plant Physiology and Biochemistry, 45(1): 1-5. DOI:10.1016/j.plaphy.2006.12.006 |
Gao M J, Allard G, Flanagan A M, et al. 2002. Regulation and characterization of four CBF transcription factors from Brassica napus[J]. Plant Molecular Biology, 49(5): 459-471. DOI:10.1023/A:1015570308704 |
Gilmour S J, Sebolt A M, Salazar M P, et al. 2000. Overexpression of Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation[J]. Plant Physiol, 124(4): 1854-1865. DOI:10.1104/pp.124.4.1854 |
Gilmour S J, Zarka D G, Stockinger E J, et al. 1998. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression[J]. Plant J, 16(4): 433-442. DOI:10.1046/j.1365-313x.1998.00310.x |
Gupta K, Agarwal P K, Reddy M K, et al. 2010. SbDREB2A, an A-2 type DREB transcription factor from extreme halophyte Salicornia brachiata confers abiotic stress tolerance in Escherichia coli[J]. Plant Cell Rep, 29(10): 1131-1137. DOI:10.1007/s00299-010-0896-7 |
Haake V, Cook D, Riechmann J L, et al. 2002. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis[J]. Plant Physiol, 130(2): 639-648. DOI:10.1104/pp.006478 |
Hong J P, Kim W T. 2005. Isolation and functional characterization of the Ca-DREBLP1 gene encoding a dehydration-responsive element binding-factor-like protein in hot pepper (Capsicum annuum L.cv Pukang)[J]. Planta, 220(6): 875-888. DOI:10.1007/s00425-004-1412-5 |
Hsieh T H, Lee J T, Charng Y Y, et al. 2002a. Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress[J]. Plant Physiol, 130(2): 618-626. DOI:10.1104/pp.006783 |
Hsieh T H, Lee J T, Yang P T, et al. 2002b. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato[J]. Plant Physiol, 129(3): 1086-1094. DOI:10.1104/pp.003442 |
Huang B, Liu J Y. 2006a. A cotton dehydration responsive element binding protein functions as a transcriptional repressor of DRE2 mediated gene expression[J]. Biochemical and Biophysical Research Communications, 343(4): 1023-1031. DOI:10.1016/j.bbrc.2006.03.016 |
Huang B, Liu J Y. 2006b. Cloning and functional analysis of the novel gene GhDBP3 encoding a DRE-binding transcription factor from Gossypium hirsutum[J]. Biochemistry and Biophysics Acta, 1759(6): 263-269. |
Huang B, Jin L G, Liu J Y. 2007. Molecular cloning and functional characterization of a DREB1/CBF-like gene(GhDREB1L)from cotton[J]. Sci China Ser C-Life, 50(1): 7-14. |
Ito Y, Katsura K, Maruyama K, et al. 2006. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold responsive gene expression in transgenic rice[J]. Plant Cell Physiol, 47(1): 141-153. DOI:10.1093/pcp/pci230 |
Jaglo K R, Gilmour S J, Zarka D G, et al. 1998. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance[J]. Science, 280(5360): 104-106. DOI:10.1126/science.280.5360.104 |
Jaglo K R, Kleff S, Amundsen K L, et al. 2001. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species[J]. Plant Physiol, 127(3): 910-917. DOI:10.1104/pp.010548 |
James V A, Neibaur I, Altpeter F. 2008. Stress inducible expression of the DREB1A transcription factor from xeric, Hordeum spontaneum L.in turf and forage grass(Paspalum notatum Flugge)enhances abiotic stress tolerance[J]. Transgenic Res, 17(1): 93-104. DOI:10.1007/s11248-007-9086-y |
Jin Taicheng, Chang Qing, Li Wangfeng, et al. 2010. Stress-inducible expression of GmDREB1 conferred salt tolerance in transgenic alfalfa[J]. Plant Cell Tiss Organ Cult, 100(2): 219-227. DOI:10.1007/s11240-009-9628-5 |
Kasuga M, Liu Q, Miura S, et al. 1999. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor[J]. Nat Biotechnol, 17(3): 287-291. DOI:10.1038/7036 |
Kasuga M, Miura S, Shinozaki K, et al. 2004. A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought-and low-temperature stress tolerance in tobacco by gene transfer[J]. Plant Cell Physiol, 45(3): 346-350. DOI:10.1093/pcp/pch037 |
Kim Y H, Yang K S, Ryu S H, et al. 2008. Molecular characterization of a cDNA encoding DRE-binding transcription factor from dehydration treated fibrous root of sweet potato[J]. Plant Physiology and Biochemistry, 46(2): 196-204. DOI:10.1016/j.plaphy.2007.09.012 |
Kizis D, Pages M. 2002. Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought responsive element in an ABA-dependent pathway[J]. Plant Journal, 30(6): 679-689. DOI:10.1046/j.1365-313X.2002.01325.x |
Kobayashi F, Ishibashi M, Takumi S. 2008. Transcriptional activation of Cor/Lea genes and increase in abiotic stress tolerance through expression of a wheat DREB2 homolog in transgenic tobacco[J]. Transgenic Res, 17(5): 755-767. DOI:10.1007/s11248-007-9158-z |
Kume S, Kobayashi F, Ishibashi M, et al. 2005. Differential and coordinated expression of Cbf and Cor/Lea genes during long-term cold acclimation in two wheat cultivars showing distinct levels of freezing tolerance[J]. Genes & Genetic Systems, 80(3): 185-197. |
Lee H J, Xiong L M, Gong Z Z, et al. 2001. The Arabidopsis HOS1 gene negatively regulates cold signal transcription and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning[J]. Genes Dev, 15(7): 912-924. DOI:10.1101/gad.866801 |
Lee J T, Prasad V, Yang P T, et al. 2003. Expression of Arabidopsis CBF1 regulated by an ABA /stress inducible promoter in transgenic tomato confers stress tolerance without affecting yield[J]. Plant Cell and Enviroment, 26(7): 1181-1190. DOI:10.1046/j.1365-3040.2003.01048.x |
Lee S C, Huh K W, An K, et al. 2004. Ectopic expression of a cold-inducible transcription factor CBF1/DREB1B in transgenic rice(Oryza sativa L.)[J]. Mol Cells, 18(1): 107-114. |
Li X P, Tian A G, Luo G Z, et al. 2005. Soybean DRE-binding transcription factors that are responsive to abiotic stresses[J]. Theor Appl Genet, 110(8): 1355-1362. DOI:10.1007/s00122-004-1867-6 |
Liu Liqing, Zhu Kai, Yang Yanfang, et al. 2008. Molecular cloning, expression profiling and trans-activation property studies of a DREB2-like gene from chrysanthemum(Dendranthema vestitum)[J]. J Plant Res, 121(2): 215-226. DOI:10.1007/s10265-007-0140-x |
Liu N, Zhong N Q, Wang G L, et al. 2007. Cloning and functional characterization of PpDBF1 gene encoding a DRE-binding transcription factor from Physcomitrella patens[J]. Planta, 226(4): 827-838. DOI:10.1007/s00425-007-0529-8 |
Liu Q, Kasuga M, Sakuma Y, et al. 1998. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis[J]. Plant Cell, 10(8): 1391-1406. DOI:10.1105/tpc.10.8.1391 |
Magome H, Yamaguchi S, Hanada A, et al. 2004. Dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor[J]. Plant J, 37(5): 720-729. DOI:10.1111/tpj.2004.37.issue-5 |
Medina J, Bargues M, Terol J, et al. 1999. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration[J]. Plant Physiol, 119(2): 463-469. DOI:10.1104/pp.119.2.463 |
Novillo F, Alonso J M, Ecker J R, et al. 2004. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis[J]. Proc Natl Acad Sci USA, 101(11): 3985-3990. DOI:10.1073/pnas.0303029101 |
Oh S J, Song S I, Kim Y S, et al. 2005. Arabidopsis CBF3/DREB1A and CBF3 in transgenic rice increased tolerance to abiotic stress without stunting growth[J]. Plant Physiology, 138(1): 341-351. DOI:10.1104/pp.104.059147 |
Owens C T, Thomashow M F, Hancock J F, et al. 2002. CBF1 orthologs in sour cherry and strawberry and the heterologous expression of CBF1 in strawberry[J]. Journal of the American Society for Horticultural Science, 127(4): 489-494. |
Park J M, Park C J, Lee S B, et al. 2001. Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco[J]. Plant Cell, 13(5): 1035-1046. DOI:10.1105/tpc.13.5.1035 |
Pellegrineschi A, Reynolds M, Pacheco M, et al. 2004. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions[J]. Genome, 47(3): 493-500. DOI:10.1139/g03-140 |
Qin F, Sakuma Y, Li J, et al. 2004. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold responsive gene expression in Zea mays L[J]. Plant and Cell Physiology, 45(8): 1042-1052. DOI:10.1093/pcp/pch118 |
Qin F, Kakimoto M, Sakuma Y, et al. 2007. Regulation and functional analysis of ZmDREB2A in response to drought and heat stress in Zea mays L[J]. Plant J, 50(1): 54-69. DOI:10.1111/j.1365-313X.2007.03034.x |
Sakuma Y, Liu Q, Dubouzet J G, et al. 2002. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREB's, transcription factors involved in dehydration and cold-inducible gene expression[J]. Biochem Biophys Res Commun, 290(3): 998-1009. DOI:10.1006/bbrc.2001.6299 |
Sakuma Y, Maruyama K, Osakabe Y, et al. 2006. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression[J]. Plant Cell, 18(5): 1292-1309. DOI:10.1105/tpc.105.035881 |
Savitch L V, Allard G, Seki M, et al. 2005. The effect of overexpression of two Brassica CBF/DREB1-like transcription factors on photosynthetic capacity and freezing tolerance in Brassica napus[J]. Plant Cell Physiol, 46(9): 1525-1539. DOI:10.1093/pcp/pci165 |
Shan D P, Huang J G, Yang Y T, et al. 2007. Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid[J]. New Phytologist, 176(1): 70-81. DOI:10.1111/nph.2007.176.issue-1 |
Shen Y G, Zhang W K, He S J, et al. 2003a. An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress[J]. Theor Appl Genet, 106(5): 923-930. DOI:10.1007/s00122-002-1131-x |
Shen Y G, Zhang W K, Yan D Q, et al. 2003b. Characterization of a DRE-binding transcription factor from a halophyte Atriplex hortensis[J]. Theor Appl Genet, 107(1): 155-161. DOI:10.1007/s00122-003-1226-z |
Stockinger E J, Gilmour S J, Thomashow M F. 1997. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit[J]. Proc Natl Acad Sci USA, 94(3): 1035-1040. DOI:10.1073/pnas.94.3.1035 |
Wang C T, Yang Q, Wang C T. 2010a. Isolation and functional characterization of ZmDBP2 encoding a dehydration-responsive element-binding protein in Zea mays. Plant Mol Biol Rep, http://springer.lib.tsinghua.edu.cn/content/5176237424881n83/Published online:28 April 2010.
|
Wang H L, Tao J J, He L G, et al. 2009. cDNA cloning and expression analysis of a Poncirus trifoliata CBF gene[J]. Biologia Plantarum, 53(4): 625-630. DOI:10.1007/s10535-009-0114-z |
Wang Xuemin, Dong Jie, Liu Yun, et al. 2010b. A novel dehydration-responsive element-binding protein from Caragana korshinskii is involved in the response to multiple abiotic stresses and enhances stress tolerance in transgenic tobacco[J]. Plant Mol Biol Rep, 28(4): 664-675. DOI:10.1007/s11105-010-0196-y |
Wang Q J, Xu K Y, Tong Z G, et al. 2010c. Characterization of a new dehydration responsive element binding factor in central arctic cowberry[J]. Plant Cell Tiss Organ Cult, 101(2): 211-219. DOI:10.1007/s11240-010-9678-8 |
Wang Yangmeng, He Congfen. 2007. Isolation and characterization of a cold-induced DREB gene from Aloe vera L[J]. Plant Mol Biol Rep, 25(3/4): 121-132. |
Wang Q, Guan Y, Wu Y, et al. 2008. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice[J]. Plant Mol Biol, 67(6): 589-602. DOI:10.1007/s11103-008-9340-6 |
Wei G, Pan Y, Lei J, et al. 2005. Molecular cloning, phylogenetic analysic expressional profiling and in vitro studies of TINY2 from Arabidopsis thaliana[J]. Journal of Biochemistry and Molecular Biology, 38(5): 440-446. |
Xiong Yanwen, Fei Shuizhang. 2006. Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.)[J]. Planta, 224(4): 878-888. DOI:10.1007/s00425-006-0273-5 |
Xue G P, Loveridge C W. 2004. HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators interacting preferably with a CT-rich element[J]. Plant J, 37(3): 326-339. DOI:10.1046/j.1365-313X.2003.01963.x |
Yamaguchi K, Shinozaki K. 2006. Transcriptional regulatory networks in cellular response and the tolerance to dehydration and cold stresses[J]. The Annual Review of Plant Biology, 57: 781-803. DOI:10.1146/annurev.arplant.57.032905.105444 |
Yang Y F, Wu J, Zhu K, et al. 2009. Identification and characterization of two chrysanthemum(Dendranthema ×morifolium)DREB genes, belonging to the AP2/EREBP family[J]. Mol Biol Rep, 36(1): 71-81. DOI:10.1007/s11033-007-9153-8 |
Zhang S J, Li N, Gao F, et al. 2010. Over-of TsCBF1 gene confers improved drought tolerance in transgenic maize[J]. Mol Breeding, 26(3): 455-465. DOI:10.1007/s11032-009-9385-5 |
Zhao H, Bughrara S S. 2008. Isolation and characterization of cold-regulated transcriptional activator LpCBF3 gene from perennial ryegrass(Lolium perenne L.)[J]. Mol Genet Genomics, 279(6): 585-594. DOI:10.1007/s00438-008-0335-4 |
Zhao Junsheng, Ren Wei, Zhi Daying, et al. 2007. Arabidopsis DREB1A/CBF3 bestowed transgenic tall fescue increased tolerance to dreought stress[J]. Plant Cell Rep, 26(9): 1521-1528. DOI:10.1007/s00299-007-0362-3 |
 2011, Vol. 47
2011, Vol. 47


