文章信息
- Su Hongyan, Wang Lei, Wang Zhongli, Zhu Luying, Kong Dongrui
- 宿红艳, 王磊, 王仲礼, 朱路英, 孔冬瑞
- Characterization and Expression of PeMYBF1 Encoding a R2R3-MYB Transcription Factor from Populus euramericana
- 欧美杨R2R3-MYB家族新基因PeMYBF1的克隆及表达
- Scientia Silvae Sinicae, 2011, 47(1): 42-49.
- 林业科学, 2011, 47(1): 42-49.
- DOI:
-
文章历史
- Received date: 2009-12-11
- Revised date: 2010-12-13
-
作者相关文章
One of the greatest challenges in plant developmental biology is to explore the genetic mechanisms underlying the great variety of floral forms. Since the tree life form imposes several different physiological and morphological constraints compared with those of herbaceous plants, the need for understanding molecular mechanisms involved in tree development has therefore become evident. However, the research in this area has been severely restricted by the large size, long generation time, and limited molecular genetic knowledge base of trees (Brunner et al., 2004; Sterky et al., 2004). Poplar (Populus spp.) has been internationally accepted as an important model tree species due to its ease of transformation, rapid growth, vegetative propagation, modest genome size and extensive expressed sequence tags (Bradshaw et al., 2000; Brunner et al., 2004; Sterky et al., 2004). Thus, studying poplar floral developmental genetics will enable new insights into the regulatory networks controlling tree development.
Given the central role of transcription factors in the regulation of development, the identification of flower-specific genes encoding transcription factors from poplar is an important step toward understanding the regulatory mechanisms underlying its flower development. The MYB gene family represents the largest transcription factor gene family in plants (Kranz et al., 1998; Riechmann et al., 2000; Stracke et al., 2001). All MYB proteins are characterized by a conserved MYB DNA-binding domain in N-terminal. This domain generally comprises up one to three imperfect repeats of 50-53 amino acids with a predicted helix-turn-helix motif. Within each repeat there are three conserved tryptophan residues spaced by 19 or 18 amino acids. Depending on the number of adjacent repeats in the MYB domain (one, two or three), MYB proteins can be classified into three subfamilies: R2R3-MYB with two repeats, MYB1R with one and MYB3R with three (Kranz et al., 1998; Stracke et al., 2001). Among them, R2R3-MYB proteins predominate in plant kingdom, and there are approximately 126 individual members in Arabidopsis alone (Riechmann et al., 2000; Stracke et al., 2001; Chen et al., 2006). Attempts to classify R2R3-MYB proteins revealed 22 distinct subgroups (Kranz et al., 1998; Stracke et al., 2001) and proteins within subgroups share homologous functions (Jin et al., 1999; Lee et al., 2001). MYB genes have been shown to be involved in diverse processes, including identity and fate of plant cells, disease resistance, response to hormones and environmental stimuli, and the regulation of phenylpropanoid metabolism, although no functional data are available for most members of this gene family (Stracke et al., 2001).
Many MYB genes are found to play important roles in flower development. For example, two members of subgroup 19, AtMYB21 and AtMYB24, have been characterized as flower-specific genes in Arabidopsis and play overlapping roles in the coordination of floral organ development, especially in anther development (Shin et al., 2002; Mandaokar et al., 2006; Yang et al., 2007). Besides, some other members of subgroup 19, including PsMYB26 from Pisum sativum, AmMYB305 and AmMYB340 from Antirrhinum majus, have been shown to be expressed primarily in flowers (Jackson et al., 1991; Uimari et al., 1997). To date, however, related research focused on some herbaceous species, such as Arabidopsis, Antirrhinum majus, Nicotiana tabacum, Zea mays, etc. In contrast, relatively few studies have been reported on the role of MYB transcription factors in transcriptional regulation of flower development in tree species. Characterization of counterparts of these MYB members from poplar, which share both high sequence similarity and expression profiles, would be an ideal starting point in the dissection of flower development in poplar (Wilkins et al., 2009).
In this study, we presented the identification of a R2R3-MYB gene, PeMYBF1, from male inflorescence of Populus euramericana. Phylogenetic analysis based on the amino acid alignment of PeMYBF1 and other related MYB proteins revealed that PeMYBF1 belonged to subgroup 19. Organ-specific expression analysis revealed that PeMYBF1 was predominantly expressed in both male and female inflorescence. It was further found that PeMYBF1 expression was tightly regulated during inflorescence development. This is the first report of the characterization and expression analysis of MYB genes involved in inflorescence development of poplar.
1 Methods and materials 1.1 Plant materialsThe adult Populus euramericana trees were cultivated in Yantai, Shandong Province, China(37°31′16″N, 121°21′24″E). Inflorescences at 5 different developmental stages were collected on 18 July, 20 August, 20 October, 2007 and 12 February, 10 April, 2008, respectively. All samples were immediately frozen in liquid nitrogen and stored at -70 ℃ until use.
1.2 Isolation of PeMYBF1 by reverse transcription PCR (RT-PCR)Total RNA was isolated from male inflorescences harvested in February, 2007 using total RNA isolation system (Promega, USA), and then was used for reverse transcriptase polymerase chain reaction (RT-PCR). A pair of degenerate primers was designed on the basis of the conserved regions of the plant R2R3-MYB protein sequences (KSCRLRW and PGRTDNE). A 167-bp fragment was amplified from cDNA by the degenerate primers. RT-PCR was carried out as described previously (Zhao et al., 2006). Comparison with sequences in the databases revealed that this fragment share the highest homology with plant R2R3-MYB genes. To isolate the 3′ and 5′ region of this gene, 3′ and 5′ RACE PCR were performed using SMARTTM RACE cDNA Amplification Kit (Clontech, USA). For 3′ RACE PCR, one specific primer:5′-GACAGCTCTTGATCATGGAAC-3′ was designed. Based on the sequence of 3′ region, a gene-specific primer:5′-CGGAGAGGTAGCTATAGACAAACC-3′ was designed for 5′ RACE PCR. Finally, the full-length cDNA was obtained by PCR amplification using the following primers: 5′ specific primer: 5′-ATGGATAAAAGTCCA TGCAAC-3′ and 3′ specific primer: 5′-CTGTCAAG ATCATTTGCCATG-3′. The PCR thermal cycles were carried out as follows: 3 min at 95 ℃, 35 cycles of 1 min at 94 ℃, 1 min at 56 ℃ and 1 min at 72 ℃, and a final extension for 10 min at 72 ℃.
PCR fragments were cloned into pGEM-T Easy Vectors (Promega, USA), introduced into Escherichia coli, and sequenced with ABI PRISMTM 377 DNA sequencer. This gene was designated as PeMYBF1 and has been submitted to GenBank with the accession No. FJ497053.
1.3 Sequence comparison and phylogenetic analysisSequence similarity between PeMYBF1 and other MYB proteins was analyzed using the blast analysis facilities at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nml.gov/). Multiple protein sequences were aligned using the Clustal W method in DNAMAN software package. The phylogenetic tree was constructed by the neighbor-joining method of MEGA 3.1 using 1 000 bootstrap replicates (
The accession No. of MYB genes analyzed are as follows: Arabidopsis thaliana AtMYB2 (NM_130287), AtMYB21 (NP_189418), AtMYB24 (AAM63674), AtMYB33 (NM_001085068), AtMYB57 (NP_186802), AtMYB62 (NM_105503), AtMYB65 (NM_111977), AtMYB78 (NM_124340), AtMYB81 (NM_128253), AtMYB101 (NM_128805); Antirrhinum majus AmMYB305 (P81391) and AmMYB340 (P81396); Petunia hybrida PhMYB3 (Z13998); Nicotiana tabacum NtMYBAS1 (AF198499) and NtMYBAS2 (AF198498); Pisum sativum PsMYB26 (CAA71992); Vitis vinifera VvMYB24 (ABW34394); Populus euramericana PeMYBF1 (FJ497053); Homo sapiens c-MYB (M15024).
1.4 Analysis of PeMYBF1 expression by RT-PCRTotal RNAs were prepared with RNeasy Plant Mini Kit (Qiagen, Germany) from various tissues, and were treated with RNase-free DNase (Promega, USA) to remove genomic DNA. RT-PCR was carried out with 2 μg of total RNA using OneStep RT-PCR Kit (Qiagen, Germany), following the manufacturer’s instructions. The primers used in the amplifications of PeMYBF1 were as follows: forward primer PeMYBF1-f2: 5′-GAGACCTATTGTCCACCATTC-3′ and reverse primer PeMYBF1-r1: 5′-CTGTCAAGATCATTTGCC ATG-3′. RT-PCR was performed as follows: 30 min at 50 ℃, 15 min at 95 ℃, 35 cycles of 1 min at 94 ℃, 1 min at 55 ℃ and 1 min at 72 ℃, and a final extension for 10 min at 72 ℃. Normalization was carried out by amplification of actin mRNA using a forward primer: 5′-ACCACATACAACTCCATCATG-3′ and a reverse primer: 5′-CACCTTGATTTCCATG CTGC-3′, under the same conditions described above. At the same time, a negative control without RNA template was also run.
The amplification products were visualized on a 1% (w/v) agarose gel via ethidium bromide staining and were digitized by estimation of the optical densities (OD) using an image analysis system (Chemilmager 4000, Alpha InfoTech Corporation, San Leandro, CA). The relative expression level of PeMYBF1 mRNA in each tissue was determined by comparison with actin expression. The data were expressed as means±SE from three different RT-PCR experiments.
2 Results 2.1 Cloning of PeMYBF1 cDNATo study the roles of MYB factors during flower development in poplar, we isolated a R2R3-MYB gene from male inflorescence of Populus euramericana by a combined RT-PCR and RACE PCR strategy, and designated it as PeMYBF1. PeMYBF1 cDNA contains a 570-bp open reading frame encoding a 190-amino acid protein with a calculated molecular mass of 21.6 ku and a theoretical pI value of 7.8. The predicted protein is a stereotypical R2R3-MYB protein, containing two MYB repeats of 53 amino acids in the N-terminal. Within the repeats, the highly conserved tryptophan residues implicated in DNA-binding were spaced by 19 or 18 amino acids. The first tryptophan of R3 repeat in the PeMYBF1 proteins is replaced by a isoleucine, which is also found in that position of other plant MYB domains (Fig. 1).

|
Fig.1 Amino acid alignment of PeMYBF1 and other subgroup 19 MYB proteins (as classified by Kranz et al., 1998) The line above indicated the R2 and R3 MYB repeats. Asterisks corresponded to positions of conserved Trp and Ile residues. The conserved motif W-MDDIW was boxed. Dark shading with white letters and gray shading with dark letters reflect 100% and 50% sequence conservation, respectively. Gaps introduced to improve alignment are indicated by dashes. |
Initial BLAST search at NCBI revealed that PeMYBF1 shared the highest homology to VvMYB24 protein from Vitis vinifera, with 69.0% overall identity and 84.5% identity in the R2R3 DNA-binding domain. PeMYBF1 also displayed great similarity to other MYB domain proteins implicated in flower development, including two Antirrhinum majus proteins (AmMYB305 and AmMYB340), two Arabidopsis proteins (AtMYB21 and AtMYB24), and one protein from Pisum sativum (PsMYB26) (Jackson et al., 1991; Uimari et al., 1997; Mandaokar et al., 2006; Yang et al., 2007). As shown in Fig. 1, in addition to the conserved DNA-binding domain in the N-terminal, PeMYBF1 shared a conversed W-MDDIW motif in C-terminal with the proteins described above (Kranz et al., 1998; Yang et al., 2007).
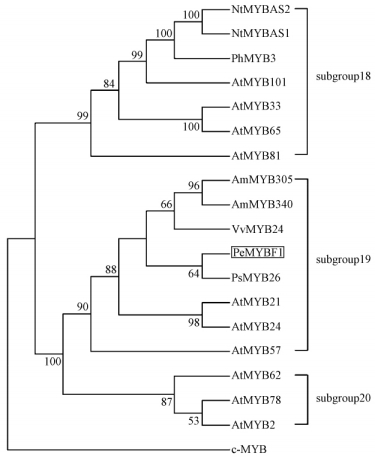
A phylogenetic tree based on amino acid alignment of PeMYBF1 and other related was constructed using the neighbor-joining method of MEGA 3.1. The dendrogram showed that PeMYBF1 belonged to the subgroup 19 (Fig. 2), together with AtMYB24, AtMYB21, AmMYB305, AmMYB340 and PsMYB26 (Kranz et al., 1998).
|
Fig.2 Phylogenetic tree of PeMYBF1 with the related MYB proteins
The neighbor-joining tree was based on the alignment of the complete coding sequences of each selected MYB proteins. The numbers indicate the bootstrap values for each node and those with < 50% support were collapsed. Subgroup18, 19 and 20 are clustered by |
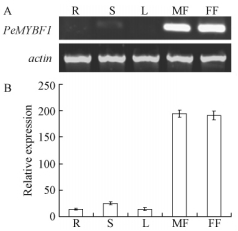
In order to examine the spatial expression pattern of PeMYBF1 in P.euramericana, total RNAs from various tissues including roots, young stems, leaves, and male and female inflorescences were analyzed by RT-PCR. An amplification product of expected size was strongly detected in male and female inflorescences, while no cDNA product was detected in roots and leaves, and little product in young stems. Actin transcript levels in these organs indicated that the amounts of total RNA in PCR reactions were similar (Fig. 3).
|
Fig.3 Organ-specific expression pattern of PeMYBF1 A. RT-PCR analysis of the expression of PeMYBF1 in different organs. B. The intensities of PCR products were measured using the image analysis system and gene expression in each organ was normalized against actin gene expression. PCR amplification of PeMYBF1 and actin were carried out for 28 cycles and 25 cycles, respectively. Data are means±SE from three different RT-PCR experiments. The standard error bar indicates the variability between repeated RT-PCR assays (n=3) from the same sample. R: Root; S: Young stem; L: Leaf; MF: Male inflorescence; FF: Female inflorescence. |
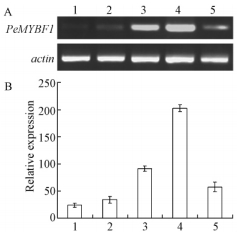
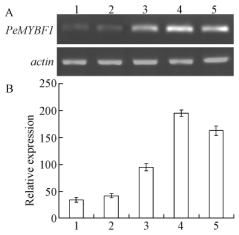
To investigate the temporal expression pattern of PeMYBF1 during inflorescence development, RT-PCR was performed on inflorescences at 5 different developmental stages which were collected on 18 July, 20 August, 20 October, 2007 and 12 February, 10 April, 2008, respectively. As shown in Fig. 4, during male inflorescence development PeMYBF1 expression was weakly detected at stage 1, and then increased with the maximum level at stage 4, whereas PeMYBF1 expression decreased significantly at stage 5. During female inflorescence development, PeMYBF1 transcripts were also weakly detectable at stage 1 and stage 2. An increase in PeMYBF1 transcripts was detected at stage 3 and the increase continued at stage 4. Different from that in male inflorescence, PeMYBF1 transcripts could still be found at high level even at stage 5 (Fig. 5).
|
Fig.4 Temporal expression pattern of PeMYBF1 during male inflorescence development A. RT-PCR analysis of the expression of PeMYBF1 in different developmental stages of male inflorescence. B. The intensities of PCR products were measured using the image analysis system and gene expression in each organ was normalized against actin gene expression. PCR amplification of PeMYBF1 and actin were carried out for 28 cycles and 25 cycles, respectively. Data are means±SE from three different RT-PCR experiments. The standard error bar indicates the variability between repeated RT-PCR assays (n=3) from the same sample. Lane 1-5, RNA from male inflorescences collected on July 18, August 20, October 20, 2007 and February 12, April 10, 2008, respectively. |
|
Fig.5 Temporal expression pattern of PeMYBF1 during female inflorescence development A. RT-PCR analysis of the expression of PeMYBF1 in different developmental stages of female inflorescence. B. The intensities of PCR products were measured using the image analysis system and gene expression in each organ was normalized against actin gene expression. PCR amplification of PeMYBF1 and actin were carried out for 28 cycles and 25 cycles, respectively. Data are means±SE from three different RT-PCR experiments. The standard error bar indicates the variability between repeated RT-PCR assays (n=3) from the same sample. Lane 1-5, RNA from female inflorescences collected on July 18, August 20, October 20, 2007 and February 12, April 10, 2008, respectively. |
The MYB family constitutes the most abundant group of transcription factors described in plants (Kranz et al., 1998; Riechmann et al., 2000; Stracke et al., 2001). Although lots of data have indicated their importance in the control of plant-specific processes, MYB genes have been much less studied in tree species than in herbaceous species (Karpinska et al., 2004; Preston et al., 2004). Recently, several members of the MYB family participating in the regulation of transcription during xylogenesis and lignin biosynthesis, which are crucial steps during wood formation, have been characterized from some tree species (Patzlaff et al., 2003a; 2003b; Karpinska et al., 2004). For instance, two R2R3-MYB genes, PtMYB1 and PtMYB4, from Pinus taeda have shown to be expressed in differentiating xylem and lignifying cells, respectively (Patzlaff et al., 2003a; 2003b). However, very little information is available on MYB genes involved in flower development of tree species.
In this study, we reported on the cloning and characterization a MYB gene, PeMYBF1, from male inflorescence of poplar for the first time. PeMYBF1 contained only two imperfect repeats 53 amino acids long in its N-terminal with high homology to other plant MYB proteins, suggesting PeMYBF1 is a putative member of R2R3-MYB gene family in poplar. When compared with all other available MYB protein sequences, PeMYBF1 showed the highest similarity to VvMYB24, a flower-specific gene of Vitis vinifera (Matus et al., 2008). Further phylogenetic analysis revealed that PeMYBF1 was located in subgroup 19 (Fig. 2), including AmMYB305 and AmMYB340 of Antirrhinum majus, and AtMYB21 and AtMYB24 of Arabidopsis, which have been shown to play important roles in flower development (Jackson et al., 1991; Uimari et al., 1997; Mandaokar et al., 2006; Yang et al., 2007). Besides, the presence of a conserved WMDDIW motif in C-terminal also supported that PeMYBF1 belonged to subgroup 19, according to the classification of MYB proteins from Arabidopsis (Kranz et al., 1998). Since classification into similar groups often reflects a functional conservation (Jin et al., 1999; Lee et al., 2001), it is reasonable to predict that PeMYBF1 is also involved in the regulation of flower development in poplar. This is consistent with the observation that PeMYBF1 transcripts accumulated predominantly in both male and female inflorescence (Fig. 3).
Further studies on PeMYBF1 temporal expression pattern revealed that during male inflorescence development PeMYBF1 transcripts abundance started at the earliest stage tested (18 July 2007), and subsequently increased until stage 4 (12 February 2008), then decreased greatly in male inflorescence at stage 5 (10 April 2008). Microscopic observation of male inflorescence showed that the perianths were already evident on 18 July. Thereafter stamens primordia appeared and by the following 12 February the microspore mother cells had entered meiosis. By 10 April, mature pollen grains were visible and some anthers dehisced (data not shown). These results suggest that the expression of PeMYBF1 is tightly related with the development of male inflorescence. PeMYBF1 expression pattern during female inflorescence development shared a similar change trend with that in male inflorescence, but transcript could still be found at a high level in mature female inflorescence (Fig. 5). To determine the temporal and spatial expression patterns within inflorescence in more detail by in situ hybridization will facilitate understanding the roles of PeMYBF1 in poplar flower development.
The most well-established role of plant MYB proteins is in the control of genes in the phenylpropanoid biosynthetic pathway (Quattrocchio et al., 1993; Franken et al., 1994; Martin et al., 1997; Yang et al., 2001). It has been reported that AmMYB305, AmMYB340, PsMYB26 regulate flavonol biosynthesis in flowers, which is one later step in phenylpropanoid metabolism and is believed to be important for pollen viability and germination (Mo et al., 1992; Taylor et al., 1992; Ylstra et al., 1994; Moyano et al., 1996; Uimari et al., 1997). Given that PeMYBF1 shared great similarity to these proteins throughout the DNA-binding domain, we assume that PeMYBF1 may also be involved in the regulation of flavonol production during poplar flower development. In order to define the precise functions of PeMYBF1,appropriate gain-of-function/loss-of-function experiments will be required to be undertaken in vivo in poplar or with a related orthologue in a plant which is more amenable to reverse genetics approaches.
Bradshaw H D, Ceulemans R, Davis J, et al. 2000. Emerging model systems in plant biology: poplar (Populus) as a model forest tree[J]. J Plant Growth Regul, 19(3): 306-313. DOI:10.1007/s003440000030 |
Brunner A M, Busov V B, Strauss S H. 2004. Poplar genome sequence: Functional genomics in an ecologically dominant plant species[J]. Trends Plant Sci, 9(1): 49-56. DOI:10.1016/j.tplants.2003.11.006 |
Chen Y H, Yang X Y, He K, et al. 2006. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family[J]. Plant Mol Biol, 60(1): 107-124. DOI:10.1007/s11103-005-2910-y |
Franken P, Schrell S, Peterson P A, et al. 1994. Molecular analysis of protein domain function encoded by the myb-homologous maize genes C1, Zm1 and Zm38[J]. Plant J, 6(1): 21-30. DOI:10.1046/j.1365-313X.1994.6010021.x |
Jackson D, Culianez-Macia F, Prescott A G, et al. 1991. Expression patterns of myb genes from Antirrhinum flowers[J]. Plant Cell, 3: 115-125. DOI:10.1105/tpc.3.2.115 |
Jin H L, Martin C. 1999. Multifunctionality and diversity within the plant MYB-gene family[J]. Plant Mol Biol, 41(5): 577-585. DOI:10.1023/A:1006319732410 |
Karpinska B, Karlsson M, Srivastava M, et al. 2004. MYB transcription factors are differentially expressed and regulated during secondary vascular tissue development in hybrid aspen[J]. Plant Mol Biol, 56(2): 255-270. DOI:10.1007/s11103-004-3354-5 |
Kranz H D, Denekamp M, Greco R, et al. 1998. Towards functional characterization of the members of the R2R3-MYB gene family from Arabidopsis thaliana[J]. Plant J, 16(2): 263-276. DOI:10.1046/j.1365-313x.1998.00278.x |
Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment[J]. Brief Bioinform, 5(2): 150-163. DOI:10.1093/bib/5.2.150 |
Lee M M, Schiefelbein J. 2001. Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis[J]. Development, 128(9): 1539-1546. |
Mandaokar A, Thines B, Shin B, et al. 2006. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling[J]. Plant J, 46(6): 984-1008. DOI:10.1111/tpj.2006.46.issue-6 |
Martin C, Paz-Ares J. 1997. MYB transcription factors in plants[J]. Trends Genet, 13(2): 67-73. DOI:10.1016/S0168-9525(96)10049-4 |
Matus J T, Aquea F, Arce-Johnson P. 2008. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes[J]. BMC Plant Biology, 8: 83-97. DOI:10.1186/1471-2229-8-83 |
Mo Y, Nagel C, Taylor L P. 1992. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen[J]. Proc Natl Acad Sci USA, 89(15): 7213-7217. DOI:10.1073/pnas.89.15.7213 |
Moyano E, Martinez G J, Martin C. 1996. Apparent redundancy in myb gene function provides gearing for the control of flavonoid biosynthesis in Antirrhinum flowers[J]. Plant Cell, 8(9): 1519-1532. DOI:10.1105/tpc.8.9.1519 |
Patzlaff A, McInnis S, Courtenary A, et al. 2003a. Characterisation of a pine MYB that regulates lignification[J]. Plant J, 36(6): 743-754. DOI:10.1046/j.1365-313X.2003.01916.x |
Patzlaff A, Newman L J, Dubos C, et al. 2003b. Characterisation of PtMYB1, an R2R3-MYB from pine xylem[J]. Plant Mol Biol, 53(4): 597-608. DOI:10.1023/B:PLAN.0000019066.07933.d6 |
Preston J, Wheeler J, Heazlewood J, et al. 2004. AtMYB32 is required for normal pollen development in Arabidopsis thaliana[J]. Plant J, 40(6): 979-995. DOI:10.1111/tpj.2004.40.issue-6 |
Quattrocchio F, Wing J F, Leppen H T C, et al. 1993. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes[J]. Plant Cell, 5(11): 1497-1512. DOI:10.1105/tpc.5.11.1497 |
Riechmann J L, Heard J, Martin G, et al. 2000. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes[J]. Science, 290(5499): 2105-2110. DOI:10.1126/science.290.5499.2105 |
Shin B, Choi G, Yi H, et al. 2002. AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1[J]. Plant J, 30(1): 23-32. DOI:10.1046/j.1365-313X.2002.01264.x |
Sterky F, Bhalerao R R, Unneberg P, et al. 2004. A Populus EST 110 resource for plant functional genomics[J]. Proc Natl Acad Sci USA, 101(38): 13951-13956. DOI:10.1073/pnas.0401641101 |
Stracke R, Werber M, Weisshaar B. 2001. The R2R3-MYB gene family in Arabidopsis thaliana[J]. Curr Opin Plant Biol, 4(5): 447-456. DOI:10.1016/S1369-5266(00)00199-0 |
Taylor L P, Jorgensen R. 1992. Conditional male fertility in chalcone synthase-deficient Petunia[J]. J Hered, 83(1): 11-17. DOI:10.1093/oxfordjournals.jhered.a111149 |
Uimari A, Strommer J. 1997. Myb26: a MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes[J]. Plant J, 12(6): 1273-1284. DOI:10.1046/j.1365-313x.1997.12061273.x |
Wilkins O, Nahal H, Foong J, et al. 2009. Expansion and diversification of the Populus R2R3-MYB family of transcription factors[J]. Plant Physiol, 149(2): 981-993. |
Yang S, Sweetman J P, Amirsadeghi S, et al. 2001. Novel anther-specific myb genes from tobacco as putative regulators of phenylalanine ammonia-lyase expression[J]. Plant Physiol, 126(4): 1738-1753. DOI:10.1104/pp.126.4.1738 |
Yang X Y, Li J G, Pei M, et al. 2007. Over-expression of flower-specific transcription factor gene AtMYB24 causes aberrant anther development[J]. Plant Cell Rep, 26(2): 219-228. DOI:10.1007/s00299-006-0229-z |
Ylstra E, Busscher J, Franken J, et al. 1994. Flavonols and fertilization in Petunia hybrida: Localization and mode of action during pollen tube growth[J]. Plant J, 6(2): 201-212. DOI:10.1046/j.1365-313X.1994.6020201.x |
Zhao Wei, Su Hongyan, Song Jian, et al. 2006. Ectopic expression of TaYAB1, a member of YABBY gene family in wheat, causes the partial abaxialization of the adaxial epidermises of leaves and arrests the development of shoot apical meristem in Arabidopsis[J]. Plant Science, 170(2): 364-371. DOI:10.1016/j.plantsci.2005.09.008 |
 2011, Vol. 47
2011, Vol. 47
