文章信息
- Chen Bihua
- 陈碧华
- Cloning and Sequence Analysis of Cinnamoyl-CoA Reductase Gene (CCR) of Pinus massoniana
- 马尾松肉桂酰辅酶A还原酶基因(CCR)克隆及分析
- Scientia Silvae Sinicae, 2009, 45(12): 46-53.
- 林业科学, 2009, 45(12): 46-53.
-
文章历史
- 收稿日期:2009-04-01
-
作者相关文章
Pinus massoniana is a native pioneer tree species for afforestat ion in barren hilly regions in southern China. It has rapid growth, wide adaptab ility, easy afforestation and restoration, low cost, resistance to drought and other excellent characteristics, as well as high-yield resin, long wood fibers, good industrial materials for paper-making and man-made fibers, playing an imp ortant role in China forestry production. High lignin content increases pollutio n from the paper-making industry and difficulty of forage digestion and absorpt ion rate for animals. The gene of regulating and controlling plant lignin conten t has become a hot study topic (Wei et al., 2001).
With negative regulation of lignin biosynthesis, the cinnamoyl-CoA reductase (CCR) catalyses hydroxyl cinnamic thioester CoA esters to the cor responding reduction of cinnamaldehyde, cinnamate after the alcohol dehydrogenase (CAD) and sinapic acid ethanol dehydrogenase (SAD), respectively, and cinnamaldehyde Songbai aldehyde reduction to the corresp onding alcohols, and lignin monomer biosynthesis catalyzes the final step. As the first branch in lignin synthesis pathway, CCR works as a potential control point regulating the flow of lignin to all C monomer, so its negative regulation may affect the lignin content (Chabannes et al., 2001; Xia et al., 2008; Ralph et al., 1998)。
CCR gene apart from the impact of lignin synthesis, also affects the fibril angle of wood (microfibral angle, referred to as MFA), which lead to material density and hardness variation (Hills, 2000). Pinus massoniana wood properties strongly affect its utility of paper and timber. The CCR gene study provides fundamentals for the further study of P. massoniana on correlation analysis, molecular breeding and gene mapping, expression.
At present, a variety of plants CCR gene has been cloned and positioned, and the correlation studies, such as polymorphism, lignin content, wood density and hardness, have been also achieved. In term of woody plants, the CCR gene studies of Pinus taeda, Picea abies,Betula luminifera, Leucaena leucocephala, Populus balsamifera and eucalypts (including 3 genera Eucalyptus, Corymbia and Angophora) and other tree species have be en reported (Anterola et al., 2002; Chen et al., 2006; 2007; Lacombe et al., 1997; McKinnon et al., 2005; Poke et al., 2006; 2003; Wadenbäck, 2006; Wadenbäck et al., 2008; Xue et al., 2008). Eucalypts are the major forest tree species for industrial raw materials in the world. Poke et al. (2003; 2004; 2006) represented thesis on the systematic studies of Eucalyptus CCR gene, involving molecular variation, the phylogenetic evolution, genetic recombination, variation and lignin content association studies. Chen et al. (2006; 2007) reported the studies of the interspecies variation and phylogeneti crelationships within the genus Corymbia or genus Angophora inferred by CCR gene.
Pinosylvin synthase gene of Pinus massoniana (Wang et al., 2008) and the introgressive hybridization between Pinus huang shanensis and P. massoniana (Luo et al., 2001) have been analyzed, however, previous studies have not shown any study of P. massoniana CCR gene. In this study, the cloning and sequencing of P. massoniana CCR gene were studied. P. massoniana CCR was aligned against that of Pinus taeda, Picea abies, Betula luminifera, Leucaena leucocephala, Populus balsamifera and eucalypts for the purpose of providing fundamentals for its correlation analysis of lignin content and wood traits, molecular marker-assisted selection, gene mapping, expression and the inter-species phylogenetic studies.
1 Materials and methods 1.1 Test materials and genomic DNA extractionNeedles were taken from a young tree of Pinus massoniana on the mountain of Fujian Academy of Forestry Sciences. Pest and disease free needles w ere preserved in the laboratory at -20 ℃. DNA was extracted using a modified CT AB method (Wang et al., 2002). The DNA purity and concentration were determined by agarose gel electrophoresis and UV spectrophotometer, and the DNA was then stored at -20 ℃.
Lambda DNA/HindⅢ Marker 2 and GeneRulerTM 100 bp plus DNA Ladder from Fermentas Life Sciences Company; TakaRa Ex Taq poly merase, TakaRa Ex Taq buffer and dNTPs produced by TaKaRa Biotec hnology (Dalian) Co., Ltd.; agarose, pUCm-T vector and DH5α cells provided by the Shanghai Sangon Biological Engineering Technology & Services Co. Ltd.; UNIQ -10 Column DNA Gel Purification Kit purchased from Shanghai Sangon Biological E ngineering Technology & Services Co. Ltd..
Primer design was referenced against Pinus taeda CCR mRNA sequence (GenBank accession No. AY064169), and was performed online by using Primer 3 (http://frodo.wi.mit.edu/cgi-bin/primer3/prim er3_www.cgi) (The White-head Institute). The primers were synthesized by TaKa Ra Biotechnology (Dalian) Co., Ltd. or Shanghai Sangon Biological Engineering Tech nology & Services Co., Ltd. 10 pairs of primers were as the followings: 1. CCR-1F:CAAGT CCCTCGGAGTACCAG, CCR-1R: GCATT GTTTCGCTGG TGTAA; 2. CCR-2F: AACGGGAGTCCTCTTGT CAG, CCR- 2R: TTTTAGTACACGATCCTCCATCA; 3.CCR-3F=CCR-1F, CCR-3R: CG AGTCAACCACATCACCAC; 4.CCR-4F: GCAGGTAAACAAACCGAGGA, CCR-4R: GCATTGTTTCGCTGGTGTAA; 5.CCR-5F: TC ACAGATG ATCCCGAAGAAATG, CCR-5R: ATTCTAATCCG AGG TC CTTGAGC; 6.CCR-6F: GTTGTTAACGGGAGTCCTCT, CCR-6R: TAGTACACGATCCTCCATCA; 7.CCR-7F: GGGAG TCCTCT TGTCAG, CCR-7R: ACACGATCCTCCAT CATATA; 8.CCR-8F: AACGGGAGTCCTCTTGTCAG, CCR-8R: CAATGTCAAACAATAATATCACAAACT; 9.CCR-9F: AACGGGAGTCCTCTTGTCAG, CCR-9R: TGTCAAACAATAATATCACAAACTTCT; 10.CCR-10F: AACGGGAGTCCTCTTGTCAG, CCR-10R: CCAATGTCAAACAATAATATCACAAAC. The sequencing primers were used for PCR amplification: CCR-3P: CCCAAGGAGGCAAGTGTAA A, CCR-4P: AGTTGGCATAGGTCTTAGCA, CCR-5P: A AATTGAGAATCCCAGATAC, CCR-6PP: CTTGCCAGGAT TAGTTAAGA, CCR-7PP: CACCAA GTTAGC AACAGAAT, AG74610(P1): ATTGCGTGGATCATAAGGAG, AG74611(P2): CTGTGCATATCCTTCTATCT. Forward sequencing primer: CCR-5P, CCR-7PP, AG74610 (P1); Reverse sequencing primer: CCR-3P, CCR-4P, CCR-6PP, AG74611 (P2). F=Forward, R=Reverse.
1.2 PCR amplificationA total of 6 μL DNA with 10 ng·μL-1 was used as template in a 25 μL PCR reaction mix containing 1 μL 20 μmol·L-1 forward and reverse primers respectively, 0.25 μL 5 U·μL-1 Ex Taq polymerase, 2.5 μL 10×Ex Taq buffer (Mg2+ Plus), 3 μL 2.5 μmol·L-1 dNTPs and 11.25 μL ddH2O. PCR amplification was carri ed out using an AB 2720 thermocycler with the following conditions: an initial hold of 5 min at 94 ℃, 30-32 cycles of 94 ℃ for 30 s, 58 ℃ for 1 min and 72 ℃ for 4 min, followed by a final hold at 72 ℃ for 4 min. PCR products were detected on 1% agrose gel electrophoresis, and visualized under UV light by ethidium bromide staining. The molecular weight ladder Markers were used for control.
1.3 PCR product cloning and sequencingSequencing templates were gel purified and cleaned using UNIQ-10 Column DNA Gel Purification Kit. Then the fragment was ligated into pUCm-T vector and transfo rmed to Escherichia coli DH5α competent cells. Sequencing was p erformed on an ABI 3730XL DNA Analyser.
The contig was done by using Vector NTI software. The sequences were aligned against Pinus taeda CCR gene encoding sequence (GenBank accession No. AY064169) for determining the gene structure, and reconstructed molecular phylogenetic trees with Neighbor-Joining method by MEGA4.0.2 software. Sequence information was submitted to GenBank (Accession No: EU753854).
2 Materials and methods 2.1 Pinus massoniana CCR gene PCR amplificationThe optimal amplification primers, primer pair 1 (CCR-1F and CCR-1R), was se lected for amplifying a fragment size of 4 389 bp (Fig. 1) via cloning and sequencing. The fragment includes 5'UTR, Exon Ⅰ to partial Exon Ⅴ (including introns), so the primer pair AG74610 (P1) and CCR-10R was used to amplify 803 bp including Exon Ⅴ and partial 3'UTR.
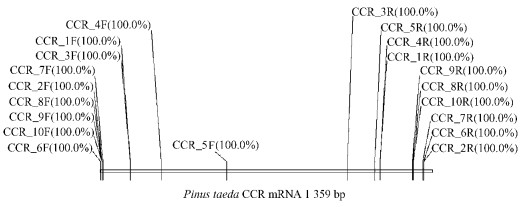
|
Fig.1 PCR amplification primer binding site diagram |
The two fragment sequences were assembled into a contig then aligned against the CCR sequences of Pinus taeda (AY064169), and Betula luminifera (FJ410450) to deduce the size and position of exons and introns. The results showed that the CCR sequence of Pinus massoniana derived was 4 530 bp (GenBank accession No. EU753854), including partial 5'UTR 121 bp, all exons, all introns, and partial 3'UTR sequence 90 bp. The sizes of Exon Ⅰ, Exon Ⅱ, Exon Ⅲ, Exon Ⅳ and Exon Ⅴ of Pinus massoniana CCR are 133, 155, 186, 353 and 148 bp, respectively; the Exon Ⅰ has the initiation codon ATG, and the Exon Ⅴ has the terminator codon TGA. One adenine nucleotide A is located on the third base upstream ATG, which matches with the “Kozak sequence" characteristics i.e. accords with the structural characteristics of the majority of eukaryotic initiation codon (i.e. ANNAUGN) (Mount, 1982). Based on these features, aligned against other species CCR sequences, the encoding nucleotides of P. massoniana CCR gene sequence was deduced to be 975 bp, encoding 324 amino acids.
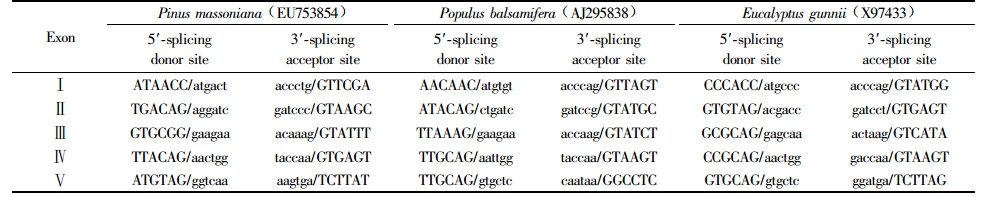
All species of genera Pinus, Eucalyptus, Corymbia, Angophora, Populus and Betula compared have equal 5 exons and 4 introns. The sizes of Exon Ⅱ, Exon Ⅲ and Exon Ⅳ are the same among the species of the six genera, but th e size variation occurs on Exon Ⅰ and Exon Ⅱ.
The junction sequences of exons and introns among P. massoniana, Populus balsamifera and Eucalyptus gunnii are not the same. The junction sequences of exon/intron of P. balsamifera and E. gunnii follow the usual rules of the genetic composition, that is, exon/GTPuAG/exon. But Intron Ⅱ/Exon Ⅲ of P. massoniana revealed as GTPuGG/Exon Ⅲ, and the rest follows the usual composition. These results indicate that the CCR gene structure of P. massoniana CCR genes is reliable (Tab. 1).
|
|
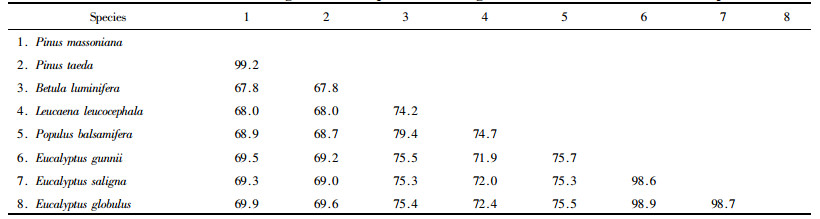
By using the biological software MEGA4.0.2 Pinus massoniana CCR gene coding region was compared with the corresponding sequence of Pinus taeda (AY064169), Populus balsamifera (AJ295838), Leucaena leucocephala (AM262869), Betula luminifera (FJ410450), Eucalyptus gunnii (X97433), Eucalyptus saligna (AF297877) and Eucalyptus globulus (AY656820), a total of 8 species (Fig. 2). The inter-species similarities were shown in Tab. 2. The results indicated that the similarities between P. massoniana and P. taeda, B. luminifera, L. leucocephala, P. balsamifera, E. gunnii, E. saligna, E. globulus were 99.2%, 67.8%, 68.0%, 68.9 %, 69.5%, 69.3%, 69.9%, respectively, of which, the similarities between P. massoniana and P. taeda is the highest, 99.2%. The similarities between P. massoniana and other broad-leaved species are lower, but at less 67.8%. Nucleotide conservation and variati on exist in these coding sequences.
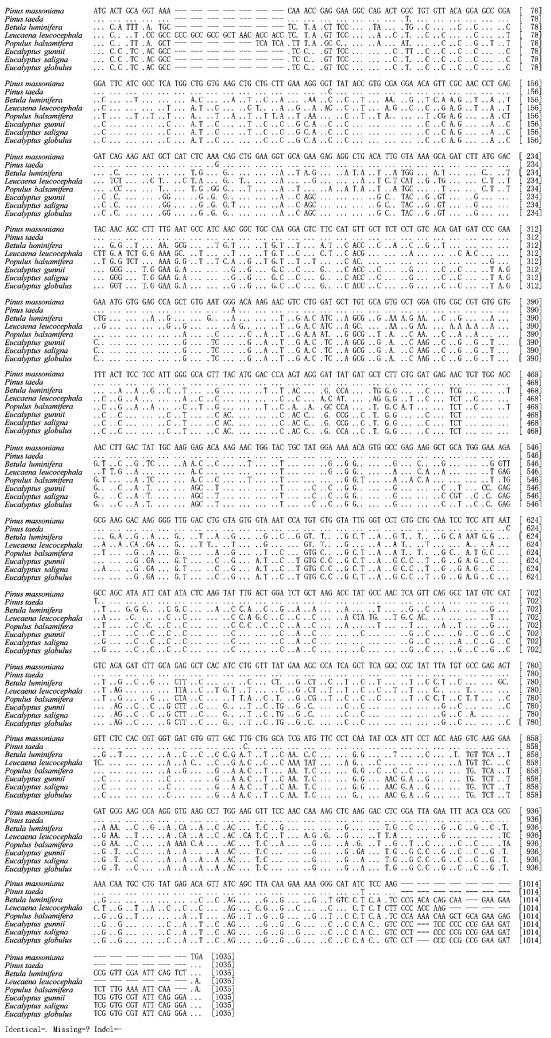
|
Fig.2 Alignment of encoding nucleotide sequences of CCR gene between Pin us massoniana and 7 species |
|
|
The results of Pinus massoniana compared with Pinus taeda at the coding nucleotide region (Fig. 2) show they have 8 conversions or transversions occurring in the coding nucleotide region 38th (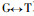
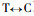

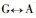

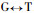
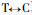
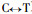
The results of Pinus massoniana compared with Betula alnoides at the coding nucleotide region indicate that they have a total of 314 conversions or transversions, in addition B. alnoides has an insertion sequence of 36 bp; Comparing P. massoniana with Leucaena leucocephala, they have a total of 312 conversions or transversions, in addition L. leucocephala has two insertion sequence, namely 21 bp and 9 bp; Comparing P. massoniana with Populus balsamifera, they have a total of 303 conversions or transversions, in addition P. balsamifera has two insertion sequence, namely 6 bp and 36 bp; Comparing P. massoniana with Eucalyptus gunnii, they have a total of 297 conversions or transversions, in addition E. gunnii has an insertion sequence of 36 bp; Comparing P. massoniana with Eucalyptus saligna, they have a total of 299 conversions or transversions, in addition E. saligna has an insertion sequence of 36 bp; Comparing P. massoniana with Eucalyptus globulus, they have a total of 293 conversions or transversions, in addition E. globulus has an insertion sequence of 36 bp.
The various origin, evolutionary history and different selection and breeding levels of different species result in the differences at DNA levels, which provides fundamentals for the studies on the species CCR origin and evolution relationship between Pinus massoniana and Pinus taeda, Betula luminifera, Leucaena leucocephala, Populus balsamifera, Eucalyptus gunnii, Eucalyptus saligna, Eucalyptus globulus.
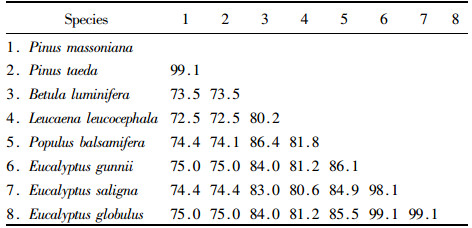
2.4 The comparative analysis of the CCR amino acid sequence of Pinus massoniana and other speciesBy using the biological software MEGA4.0.2 Pinus massoniana CCR amino acid sequence was compared with the corresponding sequence of Pinus taeda (AY064169), Populus balsamifera (AJ295838), Leucaena leucocephala (AM262869), Betula luminifera (FJ410450), Eucalyptus gunnii (X97433), Eucalyptus saligna (AF297877) and Eucalyptus globulus (AY656820), a total of 8 species (Fig. 3). The inter-species similarities were shown in Tab. 3. The results of the comparison of CCR amino acids indicate that the similarities between P. massoniana and P. taeda, B. luminifera, L. leucocephala, P. balsamifera, E. gunnii, E. saligna, E. globulus were 99.1%, 73.5%, 72.5%, 74.4%, 75.0%, 74.4%, 75.0%, respectively, of which, the similarities between P. massoniana and P. taeda is the highest, 99.1%. The similarities between P. massoniana and other broad-leaved species are lower, but at less 75.0%. Amino acid conservation and variation exist in these coding sequences.
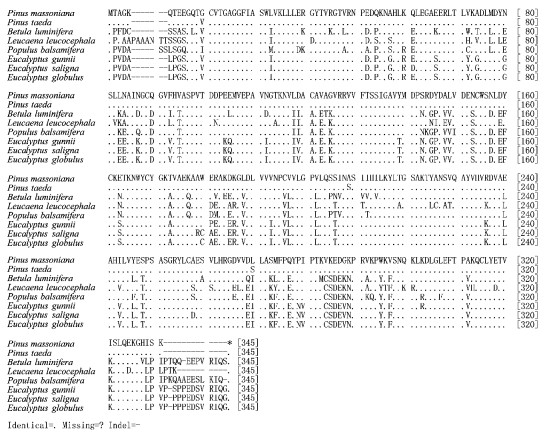
|
Fig.3 Alignment of CCR amino-acid sequences between Pinus massoniana and 7 species |
|
|
Based on the Fig. 2 and Fig. 3, comparing Pinus massoniana with Pinus taeda at the CCR encoding nucleotide, there are 8 conversions or transversions, of which 5 are synonymous mutation and 3 are non-synonymous mutation. The non-synonymous mutation occurs at the encoding nucleotide 38th (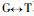
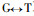
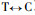

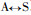
The results of Pinus massoniana compared with Betula alnoides at the protein sequence indicate that they have a total of 86 various amino acids, in addition B. alnoides has an insertion sequence of 12 amino acids; Comparing P. massoniana with Leucaena leucocephala, they have a total of 89 various amino acids, in addition L. leucocephala has two insertion sequences, namely 9 and 3 amino acids; Comparing P. massoniana with P. balsamifera, they have a total of 83 various amino acids, in addition P. balsamifera has two insertion sequences, namely 2 and 12 amino acids; Comparing P. massoniana with E. gunnii, they have a total of 81 various amino acids, in addition E. gunnii has an insertion sequence of 12 amino acids; Comparing P. massoniana with E. saligna, they have a total of 83 various amino acids, in addition E. saligna has an insertion sequence of 12 amino acids; Comparing P. massoniana with E. globulus, they have a total of 81 various amino acids, in addition E. globulus has an insertion sequence of 12 amino acids.
The various origin, evolutionary history and different selection and breeding levels of different species result in the differences at protein levels, which provides fundamentals for the studies on the species origin and evolution relations hip between Pinus massoniana and Pinus taeda, Betula luminifera, Leucaena leucocephala, Populus balsamifera, Eucalyptus gunnii, Eucalyptus saligna, Eucalyptus globulus. These provide fundamentals for the studies on these species CCR origin and evolution relationship.
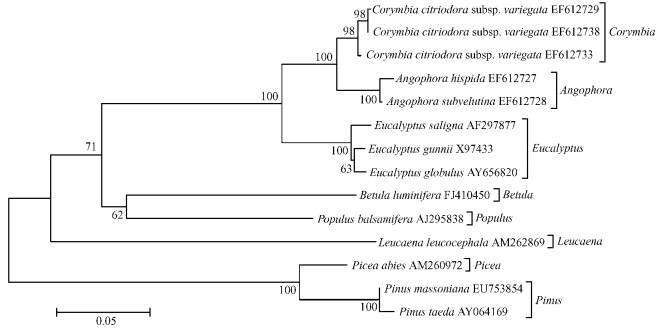
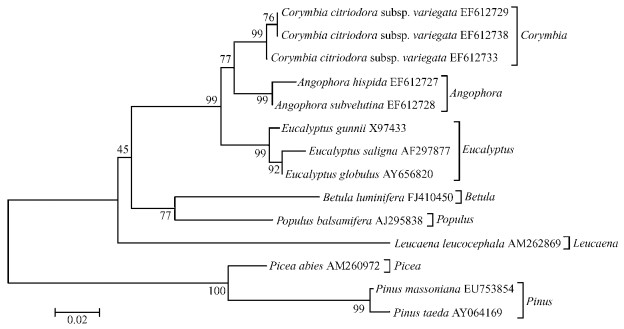
2.5 Reconstruction of phylogenetic trees using the CCR gene fragmentsThe phylogenetic trees (Fig. 4 and Fig. 5) were reconstructed by using MEGA4.0.2 with Neighbor-Joining method among P. massoniana and Pinus taeda (AY064169), Picea abies (AM260972), Populus balsamifera (AJ295838), Leucaena leucocephala (AM262869), Betula luminifera (FJ410450), Eucalyptus gunnii (X97433), Eucalyptus saligna (AF297877), Eucalyptus globulus (AY656820), Angophora subvelutina (EF612728), Angophora hispida (EF612727), Corymbia citriodora subsp. variegata (EF612729), Corymbia citriodora subsp. variegata (EF612733), Corymbia citriodora subsp. variegata (EF612738), a total of 14 tree species (or sequences), inferred from CCR gene coding nucleotide sequences (Fig. 4, Kimura 2-parameter, Bootstrap 1 000 replicates) and amino acid sequences (Fig. 5, Poisson correction, Bootstrap 1 000 replicates), respectively. The alignment sequence was from 295 bp to 879 bp of the CCR coding nucleotide sequences from P. massoniana, and the redundant sequences were removed away.
The two phylogenetic trees reflect the evolutionary relationship between P. massoniana and other species. The two phylogenetic trees reflect same genetic relationships except a little difference in bootstrap supports. The overall phylogenetic tree is divided into two clads. P. massoniana and P. taeda clusters first, then forms a clad together with Picea abies, which reveals the earliest evolution and high bootstrap, 99%-100%; Other broad-leaved tree species together form another separate clad with Leucaena leucocephala diverging earlier, followed by Populus balsamifera and Betula luminifera, and the eucalypts clusters together with high resolution among its three genera Eucalyptus, Corymbia and Angophora (45%-100% bootstrap support). Such relati onships are basically consistent with the morphology.
|
Fig.4 Molecular phylogenetic tree of Pinus masonia na and other 13 species using CCR coding nucleotide sequences |
|
Fig.5 Molecular phylog enetic tree of Pinus masoniana and other 13 species using CCR amino acid sequences |
Anterola A M, Joen J H, Davin L B, et al. 2002. Transcr iptional control of monolignol biosynthesis in Pinus taeda: fact ors affecting monolignol ratios and carbon allocation in phenylpropanoid metabolism[J]. J Biol Chem, 277: 18272-18280. DOI:10.1074/jbc.M112051200 |
Chabannes M, Barakate A, Lapierre C, et al. 2001. Strong decrease in lignin content without significant alteration of plant development is induced by simutaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants[J]. Plant J, 28: 257-270. DOI:10.1046/j.1365-313X.2001.01140.x |
Chen Bihua(陈碧华), Shepherd M, Liang Yichi(梁一池). 2006. Molecular variation of CCR gene in the spotted gums and its Corymbia relative. Journal of Southwest Forestry College(西南林学院学报), 26(5): 1-5. http://en.cnki.com.cn/Article_en/CJFDTOTAL-YNLX200605000.htm
|
Chen Bihua(陈碧华), Shepherd M, Liang Yichi(梁一池). 2007. The optimization of CCR PCR conditions, the new discovery and analysis of multi-copy gene in the genus Corymbia. Journal of Sanming University(三明学院学报), 24(2): 180-188.
|
Hills W E. 2000. Wood quality and growing to meet market requirements//IUFRO 'The Future of Eucalyptus For Wood Products', Launceston, Tasmania, Australia, 256-264.
|
Lacombe E, Hawkins S, Van Doorsselaere J, et al. 1997. Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthesis pathway: cloning, expression and phylogenetic relationships[J]. Plant J, 11: 429-441. DOI:10.1046/j.1365-313X.1997.11030429.x |
Luo Shijia(罗世家), Zou Huiyu(邹惠渝), Liang Shiwen(梁师文). 2001. Study on the introgressive hybridization between Pinus huangshanensis and P. massoniana. Scientia Silvae Sinicae(林业科学), 37(6): 118-122. http://www.linyekexue.net/EN/Y2001/V37/I6/118
|
McKinnon G E, Potts B M, Steane D A, et al. 2005. Population and phylogenetic analysis of the cinnamyol CoA reductase gene in Eucalyptus globulus (Myrtaceae)[J]. Aust J Bot, 53: 827-838. DOI:10.1071/BT04195 |
Mount S M. 1982. A catalogue of splice junction sequences[J]. Nucleic Acids Research, 10: 459-472. DOI:10.1093/nar/10.2.459 |
Poke F S, Martin D P, Steane D A, et al. 2006. The impact of intragenic recombination on phylogenetic reconstruction at the sectional level in Eucalyptus when using a single copy nuclear gene (cinnamoyl CoA reductase)[J]. Molecular Phylogenetics and Evolution, 39: 160-170. DOI:10.1016/j.ympev.2005.11.016 |
Poke F S, Vaillancourt R E, Elliott R C, et al. 2003. Sequencev ariation in two lignin biosynthesis genes, cinnamoyl CoA reductase (CCR ) and cinnamyl alcohol dehydrogenase 2 (CAD2)[J]. Molecular Breeding, 12: 107-118. DOI:10.1023/A:1026006828721 |
Poke F S, Wright J K, Raymond C A. 2004. Predicating extractives and lignin contents in Eucalyptus globulus using near infrared reflectance analysis[J]. Journal of Wood Chemistry and Technology, 24: 55-67. |
Ralph J, Hatfield R D, Piquemal J, et al. 1998. NMR characterization of altered lignins extracted from tobacco plants down-regulated for lignification enzymes cinnamyl alcohol dehydrogenase and cinnamoyl-CoA reductase[J]. Proc Nail Acad Sci USA, 95: 12803-12808. DOI:10.1073/pnas.95.22.12803 |
Wadenbäck J. 2006. Lignin studies of transgenic Norway Spruce. Ph. D. Dissertation. Swedish University of Agricultural Sciences, 1-49.
|
Wadenbäck J, Von Arnold S, Egertsdotter U, et al. 2008. Lignin biosynthesis in transgenic Norway spruce plants harboring an antisense construct for cinnamoyl CoA reductase (CCR)[J]. Transgenic Res, 17: 379-392. DOI:10.1007/s11248-007-9113-z |
Wang Bingmei(王冰梅), Guo Jinlong(郭晋隆), Ye Bingying(叶冰莹), et al. 2008. Molecular cloning and analysis of Pinosylvin Synthase gene promoter from Pinus massoniana. Subtropical Agriculture Research(亚热带农业研究), 4(4): 292-296.
|
Wang Guanglin(王关林), Fang Hongjun(方宏筠). 2002. Plant Genetic Engineering. 2nd ed. Beijing: Science Press(科学出版社), 744.
|
Wei Jianhua(魏建华), Song Yanru(宋艳茹). 2001. Recent advances in study of lignin biosynthesis and manipulation. Acta Bot Sin(植物学报), 43(8): 771-779.
|
Xia Qiongmei(夏琼梅), Duan Hongping(段红平), Tian Min(田敏), et al. 2008. Research advance in the regulation of lignin content and composition with genetic manipulation. Journal of Anhui Agricultural Science(安徽农业科学), 36(6): 2253-2256.
|
Xue Yongchang(薛永常), Zhao Yiling(赵一玲), Zhao Wenchao(赵文超), et al. 2008. Cloning and identification of lignin biosynthesis CCR gene from Populus × euramericana cv. "74/76". Journal of Liaoning Forestry Science and Technology(辽宁林业科技), (5): 18-21.
|
 2009, Vol. 45
2009, Vol. 45