文章信息
- Wang Xinrong, Zhu Xiaowei, Hu Yueqing, Huang Huanhua, Kong Xiangchao, Jia Wenhui
- 王新荣, 朱孝伟, 胡月清, 黄焕华, 孔祥超, 贾文慧
- A PCR-Based Method for Detecting Bursaphelenchus xylophilus from Monochamus alternatus
- 松墨天牛携带的松材线虫PCR检测技术
- Scientia Silvae Sinicae, 2009, 45(7): 70-75.
- 林业科学, 2009, 45(7): 70-75.
-
文章历史
- 收稿日期:2008-09-23
-
作者相关文章
2. 广东省林业科学研究院森林保护研究所 510520
2. Research Institute of Forest Protection, Forestry Academy of Guangdong Province Guangzhou 510520
Pine wilt disease caused by Bursaphelenchus xylophilus has caused serious loss to pine forestry around the world. The nematodes are transmitted by beetles. Monochamus alternatus is the most important vector of B.xylophilus (Ryss et al., 2005). It is the key point to diagnose the disease as early as possible for controlling it effectively, so the forecast of disease development trends has been studied based on the quantity of M. alternatus in the forest. A lot of research work about the relationships between the nematodes and the beetles has been done. Only adult beetles carry larvae nematodes. (Mamiya et al., 1979; Giblin-Davis et al., 1983; Kanzaki et al., 2001; Penas et al., 2006). Because nematodes are normally identified by their morphological characteristics of adult nematodes, it is difficult to identify the young nematodes carried by the beetles. A lot of molecular methods have been studied in order to identify the nematodes based on the condition that the nemtodes should be isolated from their host initially(Iwahori et al., 2000;Hoyer et al., 1998; Wu et al., 2005; Liao et al., 2001; Kang et al., 2004; Castagnone et al., 2005). As beetles usually carry several species of nematodes, it is helpful to develop the molecular method for identifying the nematodes(Wang et al., 2008). But until recently, there has been no report about the identification of nematodes from the tissue of M.alternatus based on PCR amplification.
In our study, we firstly find the proper DNA extraction method for following PCR amplification, then find one pair of B. xylophilus-specific primers for amplifying nematodes directly from the tissue of M.alternatus. Finally, a PCR-based method has been established for detecting B.xylophilus from M.alternatus without a separate DNA extraction step.
1 Materials and methods 1.1 Nematodes populations13 isolates of B. xylophilus, B.mucronatus, B.aberrans, B.corneolus, B.leoni, B.hunanensis, B.teratospicularis, Aphelenchoides resinosi, Seinura steineri, Ditylenchus parvus, Odorhabdiplogaster xiphocaudatus, Rhabditida sp. and Parasitorhabditis sp. were isolated from chips of infected pine wood of Pinus massoniana in Guangdong Province in China by the Baermann method(Xie, 2000). The nematodes species were identified under the microscope. Isolates of B.xylophilus and B.mucronatus were reared on a lawn of Pestalotiopsis sp. cultured on PDA plates, at 25 ℃ in the dark. Collected nematode species were stored in water at 4 ℃.
1.2 Genomic DNA extractionGenomic DNA of nematodes and beetle tissue were extracted, according to Zhang et al. (2001), with some modifications. 50 μL nematodes of mixed developmental stages of one species, were collected in 1.5 mL Eppendorf tube after being washed in sterilized, distilled water for 3 times, and then centrifuged at 500 g for 5 min with as much water removed as possible. Purified nematode individuals were frozen in liquid nitrogen and ground with a mortar and pestle. DNA was extracted according to a phenol/chloroform procedure. Following ethanol precipitation, DNA was re-suspended in 0.01 mol·L-1 Tris, pH 8, and 0.01 mol·L-1 EDTA and stored at -80 ℃.
DNA from single nematode of B. xylophilus, B.mucronatus, B.aberrans, B.corneolus, B.leoni, B.hunanensis, B.teratospicularis, A. resinosi, S. steineri, D. parvus, O.xiphocaudatus, Rhabditida sp., Parasitorhabditis sp., was isolated respectively based on the method of Zhang et al. (2001) by a simplified procedure: This involved a single adult or juvenile which was hand-picked and placed in 10 μL WLB(2.5 mmol·L-1 DTT, 1.125% Tween 20, 0.025% Gelatin, 125 mmol·L-1 KCl, 25 mmol·L-1 Tris-HCl(pH 8.0), 3.75 mmol·L-1 MgCl2), and placed at -70 ℃ for 30 min. Each tube was incubated at 95 ℃ for 15 min, 65 ℃ for 5 min, then 1 μL 20 mg·mL-1 proteinase K was added, with continued incubation at 65 ℃ for 1 h, and 95 ℃ for 15 min. At this stage, either the PCR reaction was performed directly in the same tube or the lysate was frozen at -20 ℃ for subsequent steps of the procedure. This procedure was modified for DNA isolation from a mixture of a single nematode + 1 mg nematode-free beetle tissues or beetle sample with B.xylophilus.
1.3 Development of species-specific PCR primer pairsTo design nucleotide sequences of species-specific PCR primer pair for B. xylophilus, DNA sequences in the ribosome region of B. xylophilus and M. alternatus had been searched from the NCBI(http://www.ncbi.nlm.nih.gov). But there is no available result for M. alternatus. Anyway, one pair of B.xylophilus-specific primers whose forward primer located in the 155th bp of ITS1 region and designated P155(sequence:5′-CTACGTGCTGTTGTTGAGTTGGC-3′) with one reverse primer located in the 538th bp of the ITS2 region and designated as P538 (sequence: 5′-TGGTGCC TAACATTGCGCGA-3′) was choosen (Fig. 1). PCR amplicon of B. xylophilus is 403 bp in the region of ribosome RNA (Hu, 2006). The primers are synthesized by Beijing SAIBAISHENG Biotic Company (Beijing, China).
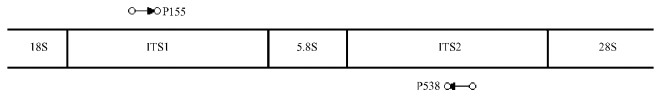
|
Fig.1 Primers location in rDNA region |
PCR amplification was performed in a total volume of 25 μL using a thermal cycle (TaKaRa). Each reaction mix contained 1.0 μL (20 ng) DNA template, 2.5 μL PCR buffer(10×), 2.5 μL MgCl2(25 mmol·L-1), 2 μL dNTP(2.5 mmol·L-1), 1.0 μL primer P155 (10 mmol·L-1)、1.0 μL primer P538 (10 mmol·L-1), 0.1 μL rTaq polymerse 0.5 U (TaKaRa Biotech, Japan) and 14.9 μL ddH2O.The cycling experimental conditions were as follows: 94 ℃ for 1 min, followed by 35 cycles at 94 ℃ for 30 s→62 ℃ for 30 s→72 ℃ for 30 s, ending with 1 cycle at 72 ℃ for 10 min and storage at 4 ℃. Amplification products were separated on a 1.5% agarose gel in 1×TAE buffer (54 mmol·L-1 Tris, 27.5 mmol·L-1 boric acid, and 10 mmol·L-1 EDTA, pH 8.0) and stained with 1 μg of ethidium bromide per mL. The band pattern was photographed under UV light. Molecular size was evaluated according to a commercial marker (TaKARA, Bio Inc., Shiga, Japan).
For amplifying DNA of a single nematode, little modification was done based on the above conditions. The cycling experimental conditions were as follows:94 ℃ for 3 min, followed by 40 cycles at 94 ℃ for 45 s→50 ℃ for 30 s→72 ℃ for 1min, and ending with 1 cycle at 72 ℃ for 10 min and storage at 4 ℃.
1.5 Assessment of the species-specificity of the primers for B. xylophilusEach of the 13 isolates of B. xylophilus, B.mucronatus, B.aberrans, B.corneolus, B.leoni, B.hunanensis, B.teratospicularis, A. resinosi, S. steineri, D. parvus, O. xiphocaudatus, R. sp., P. sp., and genomic DNA for M. alternatus was amplified with the (designated) primers respectively: Forward primer was P155 and reverse primer was P538. All amplification was performed from a single nematode. At least three replicates were performed for each treatment.
1.6 Detection of B. xylophilus from the breast gastube tissue of M. alternatusTwo sets of experiments had been done: firstly, PCR amplification was performed for one adult of B.xylophilus mixed with 2 mg nematode-free beetle tissue of M.alternatus. Six replicates were done for the treatment. Then B.xylophilus carried by the beetle tissue of M.alternatus was detected by the PCR method. Beetle breast gas tube tissue carries different numbers of nematodes (nematode number was countedbeforehand.The nematode number was from 97 to 10 860)(12 replicates). The DNA extraction procedure and PCR amplification condition was according to above-mentioned method.
2 Results 2.1 Genomic DNA extractionGenomic DNA was extracted from B.xylophilus and M.alternatus (A1/A2=1.8~1.9), which is qualified for PCR amplification (Fig. 2, 4, 5, 6). DNA from one single nematode of B.xylophilus alone (Fig. 4), or a single nematode of B.xylophilus + nematode-free beetle tissue of M.alternatus (Fig. 5), or nematodes carried by beetle tissue, of M.alternatus, carrying nematodes were also isolated for successful PCR amplification (Fig. 6).

|
Fig.2 Genomic DNA extraction result 1:Genomic DNA of B.xylophilus; 2:Genomic DNA of M.alternatus; M: DNA ladder. |

|
Fig.4 Electrophoresis of the amplicon from DNA that was isolated from 13 different nematodes species All amplifications were performed from a single individual. Four replications for each treatment had been done. B. xylophilus specific PCR products were obtained from a singlenematode of B. xylopilus (lane 1); no PCR products were obtained for any one ofthe12other nematodes species: B.mucronatus (lane 2), B.aberrans(lane 3), B.corneolus(lane 4), B.leoni(lane 5), B.hunanensis(lane 6), B.teratospicularis(lane 7), A. resinosi(lane 8), S. steineri(lane 9), D. parvus(lane 10), O. xiphocaudatus(lane 11), R. sp.(lane 12) and P. sp.(lane 13) and CK; M, DNA ladder. |
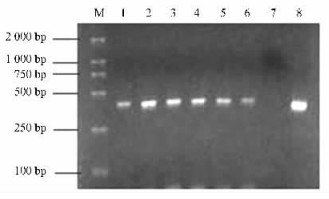
|
Fig.5 Electrophoresis of the amplification products from DNA isolated from the mixture of one single nematode, Bursaphelenchus xylophilus +2 mg beetle breast gas tube tissue Six replications for each treatment had been done. 100% B. xylophilus specific PCR products were obtained from the mixed one (lanes 1—6) and positive control (lane 8). No PCR products were obtained from the negative control (lane 7), and M, DNA ladder. |

|
Fig.6 Electrophoresis of amplicon from DNA isolated from the beetle breast gas tube tissue carrying nematodes Twelve replications for each treatment had been done. 100% B.xylophilus specific amplicons were obtained from the sample (lanes 1-12) and positive control CK+. No PCR products were obtained from the negative control CK-, and the M, DNA ladder. |
Using the forward primer of P155 and the reverse primer of P538, it was found that the expected electrophoresis band 403 bp was successfully obtained for B.xylophilus from genomic DNA (Fig. 3), and from the mixture DNA of one single nematode and nematode-free beetle tissue of M.alternatus (Fig. 4), and from the beetle tissue of M.alternatus carrying nematodes B.xylophilus (Fig. 6).

|
Fig.3 Electrophoresis of the amplicon from genomic DNA that was isolated from B. xylophilus, or from M.alternatus The forward primer was P155, the reverse primer was P538(The same below). Three replicates for each treatment. Specific amplicon of 403 bp, were obtained for B. xylophilus (lane 1) and mixed DNA of B.xylophilus and M.alternatus(lane 3). There were no amplicons for M.alternatus (lane 2) or negative control(lane 4); and M, DNA ladder. |
Each of the 13 isolates of B. xylophilus, Rhabditida sp., A. resinosi, S. steineri, D. parvus, O. xiphocaudatus, P. sp., and genomic DNA for M. alternatus were PCR amplified with the designated primers. The electrophoresis band 403 bp was only successfully obtained for B.xylophilus(Fig. 4).
2.4 Detection of B. xylophilus from the breast gastube tissue of M. alternatusThe expected amplicon of 403 bp was successfully obtained for B.xylophilus from both the mixture of one single nematode + 2 mg breast gas tube tissue of M. alternatus (Fig. 5), and beetle breast gas tube tissue with different numbers of nematodes(12 replicates)(Fig. 6).
3 DiscussionOur results show that both Genomic DNA from B.xylophilus or M.alternatus, and DNA from a single nematode of B.xylophilus alone, or single nematode of B.xylophilus + nematode-free beetle tissue of M.alternatus were isolated for successful PCR amplification. For nematodes identification, the nematode was usually isolated from the host firstly, and molecular identification was then performed (Iwahori et al., 2000; Matsunaga et al., 2004). Only Takeuchi et al. (2005) have successfully extracted nematodes DNA in infected wood by the CTAB method using phenol-chloroform procedure. In our study, we simply isolated nematodes DNA in bettle tissue by freezing and thawing them in WLB solution, which will be more convenient and healthy for identifying nematode by PCR amplification for practical use. It is the first trial in which the nematodes DNA in infected beetle tissue is isolated in this way.
A pair of B. xylophilus-specific PCR primers were choosen. The forward primer located in the 155th bp of ITS1 region was designated as P155(sequence:5′-CTACGTGCTGTTGTTGAGTTGGC-3′) with one reverse primer located in 518 th bp of ITS2 region and designated as P538 (sequence: 5′-TGGTGCCTAACATTGCGCGA-3′) (Fig. 1). PCR specific-amplicon was expected to be 403 bp (DQ855275) for B. xylophilus(Fig. 3). No amplification band was obtained for any of the other 12 nematode species related to pine wilt disease. 13 nematode species related to M.alternatus are B.xylopilus, R. sp., A. resinosi, S. steineri, D. parvus, O. xiphocaudatus, P. sp., respectively. (Fig. 4). It is the first finding that one pair of primers is effective to differentiate B. xylophilus from the 12 other nematode species related to M.alternatus according to the PCR amplification result '+/-'. It is important to continue to test the primers P155 and P538 for other nematode species related to pine wilt disease or other beetle species.
Specific PCR amplicon of 403 bp is successfully obtained by PCR amplification from the beetle tissue with B.xylophilus (Fig. 5, 6), which is the first trial to identify nematodes in beetle tissue by PCR amplification. So, it may be used in following situations: 1) Differentiating B.xylophilus from the 12 other species of nematodes as B.mucronatus, B.aberrans, B.corneolus, B.leoni, B.hunanensis, B.teratospicularis, A. resinosi, S. steineri, D. parvus, O. xiphocaudatus, R. sp. and P. sp.; 2) Detection of B.xylophilus in beetle tissue of M.alternatus, even when nematodes have already died as the result of heat, desiccation or other factors. To improve the overall usefulness of this method, further studies with more nematode species are needed.
Acknowledgements
The authors are very grateful to Dr. Esmenjaud D(INRA, France) and Dr.Eric Burt(one Canadian professor teaching English language in South China Agricultural University) for his helpful comments and linguistic corrections.
Castagnone C, Abad P, Castagnone-Sereno P. 2005. Satellite DNA-based species-specific identification of single individuals of the pinewood nematode Bursaphelenchus xylophilus(Nematoda: Aphelenchoididae). European Journal of Plant Pathology, 112: 191-193. DOI:10.1007/s10658-004-0580-2 |
Giblin-Davis R M, Kaya H K. 1983. Bursaphelenchus seanin. sp. (Nematoda: A phelenchoididae) a phoretic associate of Anthopora bomboides stanfordiana Cocker ell, 1904 (Hymenoptera: Anthoporidae). Revue de Ne′matologie, 6: 39-50. |
Giblin-Davis R M, Swan J L, Kaya H K. 1984. Bursaphelenchus kevinin. sp. (Aphelenchida: Aphelenchoididae), an associate of bees in the genus Halictus (Hymenoptera: Halictidae). Revue de Ne′matologie, 7: 77-187. |
Hu Yueqing(胡月清). 2006. Investigation and identification of nematodes in dead or dying Pinus massoniana and the detection technique of Bursaphelenchus xylophilus (枯萎松树线虫种类鉴定及松材线虫检测技术). Guangzhou, China, South China Agricultural University.
|
Hoyer U, Burgermeister W, Braasch H. 1998. Identification of Bursaphelenchus species(Nematode, Aphelenchoididae) on the basic of amplified ribosomal DNA(ITS -RFLP). Nachrichtenblatt des Deutschen Pflanzenschutzdienstes, 50(11): 273-277. |
Iwahori H, Kanzaki N, Futai K. 2000. A simple polymerase chain reaction-restriction fragment length polymorphism-aided diagnosis method for pine wilt disease. Forest Pathology, (3): 157-164. |
Iwahori H, Tsuda K, Kanzaki N, et al. 1998. PCR-RFLP and sequencing analysis of ribosomal DNA of Bursaphelenchus nematodes related to pine wilt disease. Fundam Appl Nematol, 21: 655-666. |
Liao J L, Zhang L H, Feng Z X. 2001. A reliable identification of Bursaphelenchus xylophilus by rDNA amplification. Nematologia Mediterreane, 29(1): 131-135. |
Kang J S, Choi K S, Shin S C, et al. 2004. Development of an efficient PCR-based diagnosis protocol for identification of the pinewood nematode, Bursaphelenchus xylophilus(Nematoda: Aphelenchoididae). Nematology, 6: 279-285. DOI:10.1163/1568541041217915 |
Kanzaki N, Futai K. 2001. Life history of Bursaphelenchus conicaudatus (Nematoda: Aphelenchoididae) in relation to the yellow-spotted longicorn beetle, Psacothea hilaris (Coleoptera: Cerambycidae). Nematology, 3: 473-479. DOI:10.1163/156854101753250809 |
Mamiya Y, Enda N. 1979. Bursaphelenchus mucronatus.sp. (Nematoda: Aphelenchoididae) from pine wood and its biology and pathogenicity to pine trees. Nematologica, 25: 353-361. DOI:10.1163/187529279X00091 |
Matsunaga K, Togashi K. 2004. A simple method for discriminating Bursaphelenchus xylophilus and B. mucronatus by species-specific polymerase chain reaction promer pairs. Nematology, 6: 273-277. DOI:10.1163/1568541041217960 |
Metge K, Burgermeister W G. 2005. Multiple displacement amplification of DNA for ITS-RFLP analysis of individual juveniles of Bursaphelenchus. Nematology, 7(2): 253-257. DOI:10.1163/1568541054879511 |
Penas A C, Bravo M A, Naves P, et al. 2006. Species of Bursaphelenchus Fuchs, 1937 (Nematoda: Parasitaphelenchidae) and other nematode genera associated with insects from Pinus pinaster in Portugal. Ann Appl Bio, 148: 121-131. DOI:10.1111/aab.2006.148.issue-2 |
Ryss A, Vieira P, Mota M, et al. 2005. A synopsis of the genus Bursaphelenchus Fuchs, 1937 (Aphelenchida: Parasitaphelenchidae) with keys to species. Nematology, 7: 393-458. DOI:10.1163/156854105774355581 |
Takeuchi Y, Kanzaki N, Futai K. 2005. A nested PCR-based method for detecting the pine wood nematode, Bursaphelenchus xylophilus, from pine wood. Nematology, 7(5): 775-782. DOI:10.1163/156854105775142928 |
Wang Xinrong(王新荣), Hu Yueqing(胡月清), Zhu Xiaowei(朱孝伟). 2008. Nematodes species survey and identification in pine wood of Pinus massoniana in Guangdong Province. J of Fujian College of Forestry (福建林学院学报), 28(1): 581-583.
|
Wu Xiaoqin(吴小芹), Xiong Dabin(熊大斌), An Yulin(安榆林). 2005. RAPD analysis of genetic relationships in inter and inner-species of Bursaphelenchus xylophilus and B. mucronatus. J of Nanjing Forestry University: Natural Science Edition(南京林业大学学报: 自然科学版), 29(1): 1-4.
|
Xie Hui(谢辉). 2000. Taxonomy of Plant Nematodes. Hefei: Anhui Science & Technology Press(合肥: 安徽科学技术出版社).
|
Zhang Lihai(张立海), Liao Jinling(廖金铃), Feng Zhixin(冯志新). 2001. Study on rD NA's sequence and PCR-SSCP analysis of Bursaphelenchus xylophilus. Journal of Plant Pathology(植物病理学报), 31(1): 84-89.
|
Zhao Jun(赵军). 2002. Development of computer-aided identification system of Bursaphelenchus and its Application. Master degree thesis, South China Agricultural University(华南农业大学硕士学位论文).
|
 2009, Vol. 45
2009, Vol. 45
