文章信息
- Li Mei, Shi Jisen, Li Fagen, Gan Siming
- 李梅, 施季森, 李发根, 甘四明
- Molecular Characterization of Elite Genotypes within a Second-Generation Chinese Fir (Cunninghamia lanceolata) Breeding Population Using RAPD Markers
- 杉木第2代育种群体优良基因型的RAPD遗传变异
- Scientia Silvae Sinicae, 2007, 43(12): 50-55.
- 林业科学, 2007, 43(12): 50-55.
-
文章历史
Received date: 2007-05-11
-
作者相关文章
2. 南京林业大学 国家林业局林木遗传与基因工程重点实验室 南京210037
2. Key Laboratory of Forest Tree Genetics and Gene Engineering of State Forestry Administration Nanjing Forestry University Nanjing 210037
Chinese Fir (Cunninghamia lanceolata)is a coniferous tree species belonging to the family Taxodiaceae and indigenous to China, occurring from 21°41′—33°41′N in latitude, 102°—122°E in longitude and 300 ~ 3 000 m in altitude (Chen et al., 1983).It is a diploid with n =11 chromosomes (Li et al., 1992).In natural stands it can grow impressively large, attaining 30 m in height and 3 m in girth.In many regions of China it is economically important due to its advantages in fast growth, highquality timber, and versatile end uses (Agendae Academiae Sinicae, 1978).
Cultivation of Chinese Fir is reputed back to more than 1 500 years.Nowadays, the tree accounts for approximately one quarter of China's total marketed timber productivity, and plantations of this tree species amount to more than 12.3 million hectares (Fang et al., 2005).
The economic significance of Chinese Fir has warranted active breeding programs in China since the early 1960's (Chen et al., 1987).To date, the first and second generations of breeding have been completed, and a broad set of elite genotypes have been selected as parental trees for the third breeding cycle.However, little is known about the genetic diversity of these elite genotypes as selections were made in previous generations based primarily on a limited number of morphological traits, including height, diameter at breast height (DBH) and health, with some limited selections also carried out for wood properties (Shi et al., 1993; Fang et al., 1998; He et al., 2002).
As information on diversity and relationships among elite breeding materials is of fundamental importance in plant breeding (Hallauer et al., 1988), it is imperative for the genetic diversity among the elite Chinese Fir genotypes to be characterized. Furthermore, relatively few reports have yet been published on actual measure of genetic diversity in advanced generation breeding populations for any forest tree species.
A number of methodologies can be used to efficiently quantify genetic diversity in forest tree species such as Chinese Fir. Traditionally, morphological methods have been employed to assess the genetic variation among provenances (Ye et al., 1996), half-sib families (Huang et al., 2004) and clones (Huang et al., 2005) in Chinese Fir.Further, isozymes (Mǜller-Starck et al., 1989; Yeh et al., 1994) and RAPD markers (You et al., 1998; Li et al., 2001) have been used to determine the genetic relationships among wild populations (provenances) and among elite parental genotypes.More recently, microsatellites have been used successfully for variability studies in the tree species (Chen et al., 2000).
The objective of the study was to use RAPD markers to characterize the genetic diversity and relationships among a set of 182 elite genotypes within a second-generation breeding population of Chinese Fir.
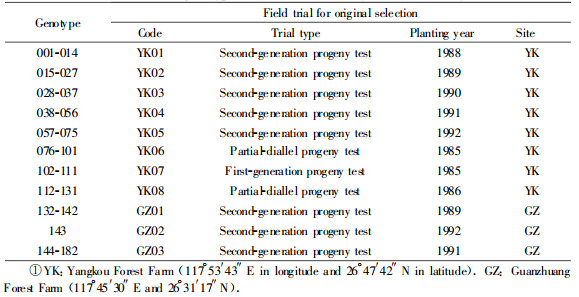
1 Materials and methods 1.1 Plant materialOne hundred and eighty-two elite genotypes of Chinese Fir within a second-generation breeding population were assessed in this study.These genotypes were selected out on basis of their superior performance in growth, stem form and wood properties from 11 field trials previously established in Fujian Province, China(Tab. 1).Each of the elite genotypes represents single openpollinated family.
|
|
Total genomic DNA was extracted from young needle tissue using the CTAB method (Doyle et al., 1990).RAPD amplification was performed in thin-walled microcentrifuge tubes using a PE9700 thermocycler (Perkin-Elmer Co.).A reaction mixture of 15 μL was composed of 1.5 μL 10 ×buffer (100 mmol·L-1 Tris-HCl pH8.3, 500 mmol·L-1 KCl, 0.01% gelatin and 5 g·L-1 BSA), 2 mmol·L-1 MgCl2, 1 unit Taq polymerase, 100 μmol·L-1 each of dATP, dTTP, dCTP and dGTP, 5 pmol 10-mer primer and 20 ng DNA template.Each reaction mixture was overlaid with 10 μL mineral oil.The amplification program included 1 min pre-denaturation at 94 ℃, then 40 cycles of 30 s denaturation at 94 ℃, 30 s annealing at 40 ℃ and 2 min extension at 72 ℃, and finally 7 min extension at 72 ℃.Amplified products were electrophoresed in 0.5 × TBE buffer through 1.2 % agarose gels, stained with ethidium bromide and photographed with a Polaroid MP4+ instant camera system.The molecular size of amplified fragments was estimated using a GeneRulerTM 100 bp Ladder (MBI).Due to concerns for the reproducibility of RAPD technology, all the RAPD reactions were performed twice to confirm the estimates obtained.
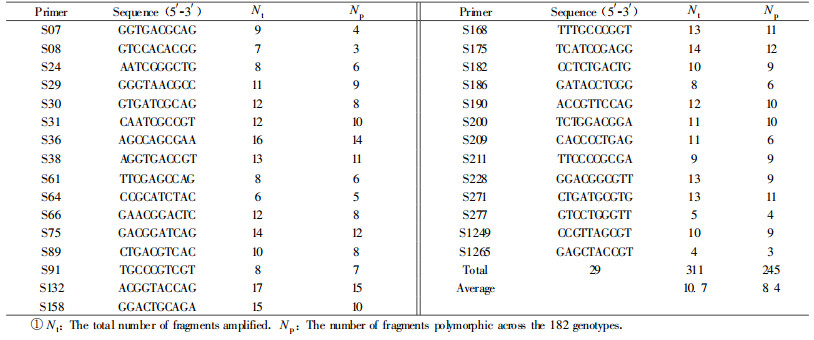
A total of 260 candidate arbitrary 10-mer primers (S01-S40, S61-S100, S121-S220, S241-S280 and S1241-S1280, Sangon Co.)were used for amplification in primer pre-screening against one DNA sample.Those that generated clear bands were then screened against 10 randomly selected genotypes.Only those primers that gave clear and polymorphic bands were applied to all the samples (Tab. 2).
|
|
RAPD markers were manually scored as "1" for presence and "0" for absence over all the genotypes studied.Only clear and reproducible bands in each of the two replications were considered for further analysis.Each marker was designated with the primer name, a hyphen " -" and band size (bp).
The RAPD data were analyzed using the computer software POPGENE 1.32 (Yeh et al., 1999).Pair-wise genetic distance between genotypes was calculated according to Nei (1978)and subsequently used to generate a dendrogram using the unweighted pair-group with arithmetic average (UPGMA)method (Sneath et al., 1973).
The analyses of molecular variance (AMOVA)were performed to certify the significance of genetic divergence among and within the field trials, except trial GZ02 that contained only one elite genotype, using the RAPD data and the software WINAMOVA 1.55 (Excoffier et al., 1992).The AMOVA procedure provides an estimate of population differentiation, Φst, analog of the F statistic, as the inter-population distance average between any two populations (Gustine et al., 1999).
2 Results 2.1 RAPD polymorphismTwenty-nine primers were finally selected out for amplification over all the 182 genotypes (Tab. 2).The 29 primers generated a total of 311 repetitive, distinct and visually clear bands, 245 (78.8%)of which were polymorphic.The molecular size of the bands ranged from 100 bp to 3 500 bp.The number of bands produced by a single primer ranged from six (primer S64)to 17 (primer S132)with a mean of 10.7.The number of polymorphic fragments ranged from three to 15 with a mean of 8.4 (Tab. 2).Meanwhile, the size of amplification products varied with primer.For instance, bands generated by primer S175 were from 300 bp to 2 700 bp in size, whereas those of primer S182 were all less than 700 bp.
2.2 Genetic relationships among elite Chinese Fir genotypesA relatively wide range of genetic distance values among genotypes was observed, with most falling between 0.3 and 0.7 (data not shown).The highest value of Nei's genetic distance observed was 0.828 4 between genotypes 013 and 098, whilst the lowest was between genotypes 013 and 014 as well as 127 and 128, each pair with a value 0.228 3.
An UPGMA dendrogram based on Nei's genetic distance allowed for 11 clusters to be distinguished over the 182 genotypes tested (Fig. 1).The first through eleventh clusters comprised of 30, 66, 20, 9, 15, 15, 2, 10, 8, 3 and 3 genotypes, respectively. In general, genotypes from the same field trial tended to fall into different clusters.Most of these clusters could be classified into three main groups at a cutting-point of 0.54 :group Ⅰ (cluster 01), group Ⅱ (clusters 02 -06) and group Ⅲ (clusters 07 - 10)(Fig. 1).The rest of the genotypes formed an independent subgroup (cluster 11).

|
Fig.1 UPGMA cluster analysis of 182 Chinese Fir elite genotypes based on Nei's genetic distance |
Analyses of the genetic diversity among and within the field trials, except trialGZ02, showed that a relatively high proportion of total genetic diversity resided within field trials (96.8 %), whereas the genetic diversity among field trials was low but significant (Φst =0.032, P < 0.001)(Tab. 3).
|
|
The RAPD technique was successfully used in this study to characterize the genetic diversity and relationships among 182 elite genotypes selected from the second-generation breeding population of Chinese Fir.High levels of RAPD polymorphism were observed, with the percentage of polymorphic loci 78.8% and indicating a high level of genetic diversity within the breeding population. Such polymorphism agrees well with results obtained in a separate study of seven Chinese Fir provenances using the same marker assay (79.8 %) (You et al., 1998). Furthermore, this polymorphism is slightly higher than that observed in a separate study of 30 superior Chinese Fir parental trees (66.9%)(Li et al., 2001), which could be mainly due to the larger sample size and hence a better representation of genotypes analyzed. However, the proportion of polymorphic loci was lower than that observed in 16 natural populations of the species (88.0%), detected with allozymes (Yeh et al., 1994).This is probably related to the nature of the populations evaluated, since breeding practices can reduce the genetic diversity available. Reduction of genetic diversity caused by breeding has been reported in crops, such as wheat (Triticum aestivum)(Roussel et al., 2004; Fu et al., 2005; 2006) and sorghum (Sorghum spp.) (Jordan et al., 1999).Alternatively, the co-dominance nature of the allozyme markers used by Yeh et al. (1994)may be attributable in respect to their capacity in detection of heterozygosity.Nevertheless, there is little evidence to support the hypothesis that genetic diversity has been significantly reduced over time in our Chinese Fir breeding program.
Although molecular diversity has been assessed in breeding populations for a number of forest tree species, e.g.Timor Mountain Gum (Eucalyptus urophylla)(Gaiotto et al., 1997; Leite et al., 2002) and Turkey Pine (Pinus brutia)(Kandedmir et al., 2004), this is the first of its kind for an advanced-generation breeding population.In the present study, high genetic diversity was revealed in breeds for advanced generation breeding in Chinese Fir, which implies that a broad genetic base is still maintained in the breeding population even after two generations of selection, and there is good potential for further genetic gains to be obtained through ongoing breeding.
Chinese Fir has the potential to be propagated en masse through various vegetative means, including rooted cuttings and tissue culture, for large scale establishment.However, identification of superior Chinese Fir clones for mass vegetative propagation cannot be done successfully on morphological traits alone, due to the availability of a limited number of morphological selection traits.Thus, there is a need for selection of candidate clones to be assisted by molecular markers.In this respect RAPD methodology can be of value for identification and commercial registration of Chinese Fir clones, as it is more reliable, efficient and direct as compared to the traditional morphological approach.On the other hand, the high polymorphisms shown by RAPD data may suggest that the selected genotypes examined in this study could be used as a suite of clones for Chinese Fir cultivation without concerns about limited genetic diversity within a plantation mixture comprising these genotypes.
The wide range of genetic distance values and the dendrogram generated indicate that the 182 genotypes studied were genetically quite diverse, and the lack of a clear common clustering of the genotypes from the same field trial also revealed a diverse origin of the material tested.In retrospect, Chinese Fir breeding programs, started in the early 1960s, mainly followed a strategy of half-sib recurrent mass selection without keeping family identity.Such a strategy has been adopted for early breeding programs in many forest tree species (Shelbourne, 1969; Boland, 1981).A significant concern about such strategies is a rapid reduction of genetic diversity that can arise after even a few generations of intense selection, which can in turn lead to inbreeding depression.The low genetic distance between each of a few pairs of Chinese Fir genotypes, e.g.013 and 014 or 127 and 128 in this study, would possibly indicate that such a pair of genotypes share common pedigree and can thereafter result in the risk of inbreeding.Efficient ways to overcome such problems include selection across field trials located in multiple environments to ensure a wide range of adaptability and also the infusion of new unrelated materials in each generation to broaden the genetic base.Such factors were the case in Chinese Fir.For instance, the elite genotypes analyzed here were selected from multiple environments, say, 11 field trials, and new unrelated genotypes were recruited, namely, genotypes 102 -111 derived from the firstgeneration progeny test (trial YK08).Further, involvement of control-pollinated materials, e.g.genotypes 076 -101 (trial YK06) and 112 -131 (trial YK08), would also have helped to efficiently constrain the gene flow within a subset of parental breeds and thereby reduce the possibility of inbreeding.
The RAPD analyses detected high genetic differentiation among the genotypes within field trials (96.80 %) and a low variability between field trials (3.20 %).This high variability within trials is similar to the variabilities that have been reported among natural populations of a number of tree species, e.g.96.86 % within Mediterranean zones in olive (Olea europaea) (Belaj et al., 2002) and 92.81% within natural populations in Monarch Birch (Betula maximowicziana)(Tsuda et al., 2005).
The low but significant level of differentiation observed in this study among the field trials (Φst =0.032, P < 0.001) indicates the presence of phenotypic differentiation in adaptability among field trials.This differentiation is probably attributed to the local selection of the genotypes in various field trials for superior performance in morphological characters.
In conclusion, high levels of genetic diversity are demonstrated within the elite genotypes for advanced generation breeding in Chinese Fir.The results are important for breeding population management and future breeding practices of this tree species.The present study represents the first to characterize the breeds for advanced-generation breeding at the molecular level in perennial forest tree species.
Agendae Academiae Sinicae (中国科学院中国植物志编辑委员会).1978.Flora Reipublicae Popularis Sinicae—Tomus 7.Beijing: Science Press(科学出版社), 284-289
|
Belaj A, Satovic Z, Rallo L, et al. 2002.Genetic diversity and relationships in olive (Olea europaea L.)germplasm collections as determinedby randomly amplified polymorphic DNA.Theor Appl Genet, 105: 638-644 https://link.springer.com/article/10.1007/s00122-002-0981-6
|
Boland D J.1981.Eucalypt seed for Indian plantations from better Australian natural seed sources.Indian For, 107: 125-134
|
Chen Bowang(陈伯望), Hong Jusheng(洪菊生), Shi Xingbo(施行博).2000.Study ongenetic diversity of Cunninghamia lanceolata and Taiwania flousiana by using chloroplast microsatellites.Sci Silvae Sin(林业科学), 36(3): 46-51 http://www.linyekexue.net/EN/abstract/abstract3951.shtml
|
Chen Yuewu(陈岳武), Shi Jisen(施季森).1983.Some fundamental problems in genetic improvement of Chinese fir.J Nanjing Tech Col For Prod(南京林产工业学院学报), 7(4): 5-19 http://en.cnki.com.cn/Article_en/CJFDTOTAL-NJLY198304001.htm
|
Chen Yuewu(陈岳武), Shi Jisen(施季森).1987.Genetic variation and improvement program of Chinese Fir.J Subtrop For Inst(亚热带林业科学研究), (1): 1-19
|
Doyle J J, Doyle D L.1990.Isolation of plant DNA from fresh tissue.Focus, 12: 13-14
|
Excoffier L, Smouse P, Quattro JM.1992.Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data.Genetics, 131: 479-491 https://www.ncbi.nlm.nih.gov/pubmed/1644282
|
Fang Lejin(方乐金), Shi Jisen(施季森), Zhang Yunbin(张运斌), et al. 1998.Studies on comprehensive selection for superior family and clone of Chinese Fir. J Nanjing For Univ(南京林业大学学报), 22(1): 17-21 http://en.cnki.com.cn/Article_en/CJFDTOTAL-NJLY801.004.htm
|
Fang Xi(方晰), Tian Dalun(田大伦), Xiang Wenhua(项文化), et al. 2005.Soil CO2 release rate and its effect factors in Chinese fir plantation.Sci Silvae Sin(林业科学), 41(2): 1-7 http://www.linyekexue.net/EN/abstract/abstract4518.shtml
|
Fu Y B, Peterson G W, Richards K W, et al. 2005.Allelic reduction and genetic shift in the Canadian hard red spring wheat germplasm released from 1845 to 2004.Theor Appl Genet, 110: 1505-1516 https://link.springer.com/article/10.1007/s00122-005-1988-6
|
Fu Y B, Peterson G W, Yu J K, et al. 2006.Impact of plant breeding on genetic diversity of the Canadian hard red spring wheat germplasm as revealed by ESTderived SSR markers.Theor Appl Genet, 112: 1239-1247 https://link.springer.com/article/10.1007/s00122-006-0225-2
|
Gaiotto F A, Bramucci M, Grattapaglia D.1997.Estimation of outcrossing in a breeding population of Eucalyptus urophylla with dominant RAPD and AFLP markers.Theor Appl Genet, 95: 842-849 https://link.springer.com/article/10.1007/s001220050634
|
Gustine D L, Huff D R.1999.Genetic variation within and among white clover populations from managed permanent pastures of the northeastern USA.Crop Sci, 39: 524-530 http://dl.sciencesocieties.org/publications/cs/abstracts/39/2/CS0390020524
|
Hallauer A R, Miranda J B.1988.Quantitative genetics in maize breeding.2nd ed.Iowa State University Press, Ames, Iowa
|
He Guiping(何贵平), Chen Yitai(陈益泰), Zhang Guowu(张国武).2002.Genetic analysis and family selection for main traits of growth and wood quality of Chinese fir.For Res(林业科学研究), 15(5): 559-563 http://en.cnki.com.cn/article_en/cjfdtotal-lykx200205009.htm
|
Huang Shouxian(黄寿先), Shi Jisen(施季森), Li Li(李力), et al. 2005.Genetic variation on tracheid microfibril angle of Chinese fir clones.J Nanjing For Univ(南京林业大学学报), 29(1): 11-14 http://en.cnki.com.cn/Article_en/CJFDTOTAL-NJLY200501002.htm
|
Huang Shouxian(黄寿先), Zhou Chuanming(周传明), Zhu Liqiong(朱栗琼), et al. 2004.Study on the genetic variation of growth traits and wood properties for Chinese fir half-sib families.Guihaia(广西植物), 24(6): 535-539 http://en.cnki.com.cn/Article_en/CJFDTOTAL-GXZW200406009.htm
|
Jordan D, Tao Y, Godwin I, et al. 1999.Loss of genetic diversity associated with selection for resistance to sorghum midge in Australian sorghum.Euphytica, 102: 1-7 https://link.springer.com/article/10.1023/A%3A1018311908636
|
Kandedmir G E, Kandedmir I, Kaya Z.2004.Genetic variation in Turkish Red Pine (Pinus brutia Ten.)seed stands as determined by RAPD Markers.Silvae Genet, 53: 169-175 https://www.researchgate.net/publication/287685383_Genetic_Variation_in_Turkish_Red_Pine_Pinus_brutia_Ten_Seed_Stands_as_Determined_by_RAPD_Markers
|
Leite S M M, Bonine C A, Mori E S, et al. 2002.Genetic variability in a breeding population of Eucalyptus urophylla S.T.Blake.Silvae Genet, 51: 5-6
|
Li Mei(李梅), Shi Jisen(施季森), He Zhenxiang(何祯祥), et al. 2001.Study on molecular genetic variation of superior trees in Chinese fir (Cunninghamia lanceolata (Lamb.)Hook).Sci Silvae Sin(林业科学), 37(4): 137-141 http://www.linyekexue.net/EN/abstract/abstract71.shtml
|
Li Zhanlin(李占林), Song Yunchun(宋运淳), Liu Lihua(刘礼华), et al. 1992.Karyotype analysis of 8 geographic seed sources of Cunninghamia lanceolata. J Central-south For Col(中南林学院学报), 12(1): 11-17
|
Mǜller-Starck G, Liu Y Q.1989.Genetics of Cunninghamia lanceolata Hook. 2. Genetic variation within and between two provenance samples.Silvae Genet, 38: 172-177
|
Nei M.1978.Estimation of average heterozygosity and genetic distance from a small number of individuals.Genetics, 89: 583-590 http://med.wanfangdata.com.cn/Paper/Detail/PeriodicalPaper_PM17248844
|
Roussel V, Koenig J, BechertM, et al. 2004.Molecular diversity in French bread wheat accessions related to temporal trends and breeding programmes.Theor Appl Genet, 108: 920-930 http://med.wanfangdata.com.cn/Paper/Detail/PeriodicalPaper_PM14614567
|
Shelbourne C J A.1969.Tree breeding methods.A review of tree breeding strategies in relation to classical plant breeding methods with quantitative genetic expectations of gain, and predictions using data from Pinus radiata. New Zealand Forest Service Technical Paper No.55
|
Shi Jisen(施季森), Ye Zhihong(叶志宏), Weng Yuzhen(翁玉榛), et al. 1993.Research on the joint genetic improvement of growth and wood properties in Chinese fir (Cunninghamia lanceolata (Lamb.)Hook).J Nanjing For Univ(南京林业大学学报), 17(1): 1-8 http://en.cnki.com.cn/Article_en/CJFDTOTAL-NJLY199301000.htm
|
Sneath P H A, Sokal R R.1973.Numerical Taxonomy.San Francisco : WH Freeman Press
|
Tsuda Y, Ide Y.2005.Wide-range analysis of genetic structure of Betula maximowicziana, a long-lived pioneer tree species and noble hardwood in the cool temperate zone of Japan.Mol Ecol, 14: 3929-3941
|
Ye Zhihong (叶志宏), Shi Jisen(施季森), He Zhenxiang(何祯祥).1996.Assessing genetic stability of Chinese fir genotypes using a spatial model.J Nanjing For Univ(南京林业大学学报), 20(2): 1-4
|
Yeh F C, Boyle T.1999.POPGENE version 1.32.The user-friendly software for population genetic analysis.University of Alberta and CIFOR, Calgary, Alta
|
Yeh F C, Shi J, Yang R, et al. 1994.Genetic diversity and multilocus associations in Cunninghamia lanceolata (Lamb.)Hook from the People's Republic of China.Theor Appl Genet, 88: 465-471 https://link.springer.com/article/10.1007/BF00223662
|
You Yong(尤勇), Hong Jusheng(洪菊生).1998.Application of RAPD marker to genetic variation of Chinese fir provenances.Sci Silvae Sin(林业科学), 34 (4): 32-38 http://www.linyekexue.net/EN/abstract/abstract2701.shtml
|
 2007, Vol. 43
2007, Vol. 43