文章信息
- Gan Siming, Shi Jisen, Bai Jiayu, Wu Kunming, Wu Juying
- 甘四明, 施季森, 白嘉雨, 吴坤明, 吴菊英
- DNA POLYMORPHISM AND HETEROZYGOSITY IN SEVEN PARENTAL EUCALYPT CLONES AS REVEALED BY RAPD MARKERS
- 7个桉树杂交亲本RAPD位点多态性和杂合性的研究
- Scientia Silvae Sinicae, 2003, 39(2): 162-167.
- 林业科学, 2003, 39(2): 162-167.
-
文章历史
Received date: 2000-04-13
-
作者相关文章
2. 南京林业大学林木遗传与基因工程实验室 南京 210037
2. Laboratory of Forestry Genetics and Gene Engineering, Nanjing Forestry University Nanjing 210037
Eucalyptus species(family Myrtaceae) are native to Australia, Indonesia and the adjacent islands and largely introduced for plantation establishment in global tropics and subtropics due to their fastness in growth, multiplicity of uses and plasticity of adaptations (Jacobs, 1981; Eldridge et al., 1993). E. urophylla S.T. Blake and E. tereticornis Smith are two of the most important species in the genus. In China, substantial genetic gain in such traits as growth and wood properties has been demonstrated in field trials through hybridization and recurrent selection in the two trees (Wu et al., 1996; Xu et al., 1996). The two species along with their hybrids are of great potential in establishing short-rotation and industry-oriented (e. g. pulp and paper making)plantations in worldwide context, and great efforts have been placed on their multiple objective improvement, especially with a focus on integrating modern biological techniques.
A successful breeding program should take full advantage of broad genetic base, for such poorly improved species as forest trees in particular, and genetic polymorphism and heterozygosity are always important indices used to monitor the genetic diversity (Lanham, 1996). Isozymes were initially sought in plant for such purpose (Hamrick et al., 1990), but their use was limited at some extent by the low amount of variability. Recent development of molecular marker technology has made it possible to conduct such studies with more powerful assays, and random amplified polymorphic DNA (RAPD)is preferred in this term because it is easy to analyze, requires very little DNA and does not need radioactivity handling facilities(Williams et al., 1990; Welsh et al., 1990; Gan et al., 1998; Martin et al., 1993). Though the reproducibility of RAPD markers are sometimes questioned, reliable results have been verified in many cases of plants (Xiao et al., 1996; Lanza et al., 1997). Now DNA polymorphism and heterozygosity have been potently identified with RAPD markers in quite a few species, such as intraspecific crossing of crop (Brassica napus)(Marshall et al., 1994), cultivars of tree (Ribes nigrum L.) (Lanham, 1996), and sibs of a single urediniospore-derived culture of fungus (Cronartium quercuum f. sp. fusiforme) (Doudrick et al., 1993).
In this study, we use RAPD markers and 5 full-sib families to detect the levels of polymorphism and heterozygosity in seven parental clones of E. urophylla S. T. Blake (3 females) and E. tereticornis Smith (4 males). The implications for material selection in linkage mapping studies are discussed.
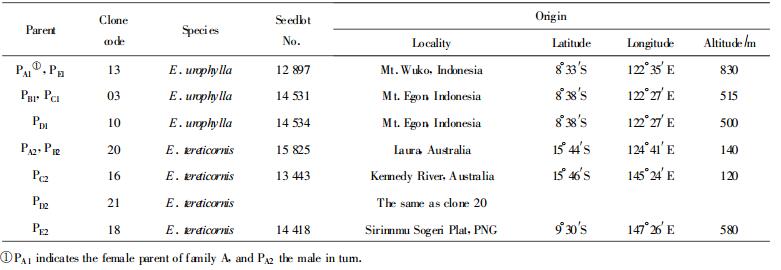
1 Materials and Methods 1.1 Plant materialsThree maternal clones of E. urophylla and four paternal ones of E. tereticornis were employed in this study, which seedlings were raised in 1989 with seeds collected in natural forests (by courtesy of CSIRO, Australia). Details of origin and code of the parental clones are shown in Table 1. The trees were conserved in the gene pool in Research Institute of Tropical Forestry, CAF, and grafted in 1992. Controlled pollination was made in 1996 to produce five full-sib families A, B, C, D and E(Table 1), and more than ten sib seedlings per family were then raised in nursery with seeds harvested. For each family, both parents and ten sibs were used for RAPD analysis.
|
|
Fresh tender leaves about 1 g per parent and 5-month-old sib seedling were sampled and stored at -20 ℃ for subsequent DNA extraction. Preparation of DNA was followed with the procedure describe in Doyle and Doyle (1990), amended by the addition of 5% polyvinylpyrrolidone (PVP) and 2% 2-mercaptoethanol to the extraction buffer. DNA concentration was estimated by electrophoresis of agarose gel stained with ethidium bromide (EB)basing on the fluorescence intensities of the sample.
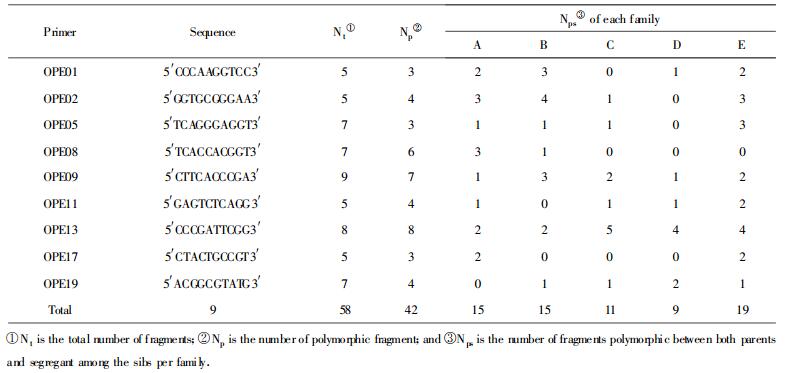
RAPD was performed in thin-walled microcentrifuge tubes with Idaho Rapidcycler (Idaho Technologies). The amplification program and RAPD reaction mixture were in accordance with Gan et al(2000). A total of 20 candidate primers (Operon Technologies Inc.), namely OPE01~OPE20, were used for primer screening against the seven parental DNA templates, and 9 primers that could produce polymorphic, clear and stable fragements were eventually selected for formal amplification (Table 2).
|
|
RAPD genotypes of 7 parents were scored for the presence absence of amplification fragments, with 1 representing presence and 0 absence of a fragment. Polymorphic markers among parents were recorded for polymorphism estimation. Genetic distance of different parent pair was then calculated according to Nei's method (1978). The number of heterozygous loci per parent was evaluated on the basis of the segregation across the sibs, and heterozygosity was calculated as the percentage of heterozygous loci taking of the total number of fragments per parental clone.
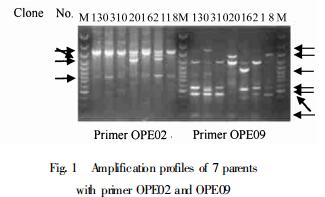
2 Results and Discussion 2.1 DNA polymorphism among parental clonesTotally 58 fragments were amplified with 9 primers selected out of 20 candidates, including 42(72. 4%)being polymorphic and 16 persistent (non-polymorphic) among the 7 parental clones (Table 2). The number of fragments varied with primer. Two unique primers OPE09 and OPE13 could discriminate clearly between all the parents. The high level of polymorphism might be indicative of a relatively high level of polymorphism for the parents studied (Nei, 1978). Figure 1 shows the RAPD profiles of 7 parents amplified with primer OPE02 and OPE09.
|
Fig.1 Amplification profiles of 7 parents with primer OPE02 and OPE09 The arrows show the polymorphic fragments. M is the 100bp DNA ladder size standard(MBI) (with fragments representing 3 000, 2 000, 1 500, 1 200, 1 031, 900, 800, 700, 600, 500, 400, 300, and 200bp from the top to the bottom) |
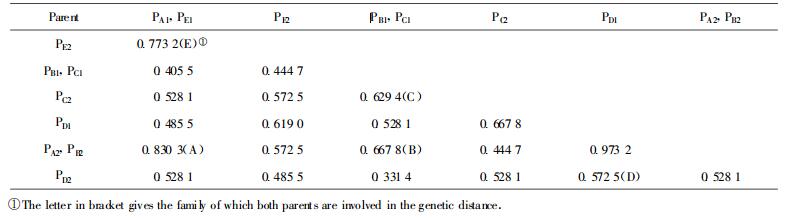
Pairwise comparisons of RAPD data between parental clones resulted in a matrix of genetic distance (GD)(Table 3). The five families followed in parental GD a descending order A>E>B>C >D. This order was roughly the same as clone fingerprints previously amplified with other primers (Gan et al., 1999).
|
|
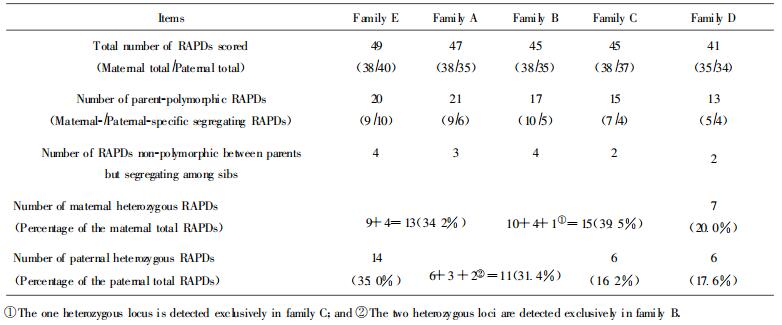
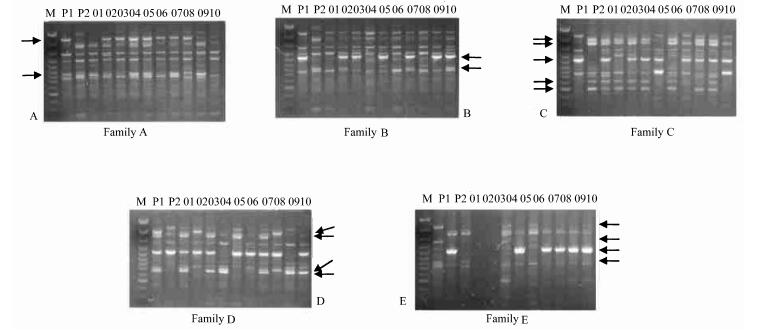
According to the dominant nature of RAPD markers, we could deduce the genotype AA or Aa at a locus from the presence of a RAPD fragment and recessive homologous aa of the absence. Thus, the fragment-specific parent could be genotyped as heterozygous Aa if the fragment segregated in the offspring in a bi-parental diploid mode. In this way the number and percentage of putative heterozygous RAPD loci (RAPDs)of each parent were calculated (Table 4). The average of heterozygosity was calculated as the percentage that the total heterzygous loci took of the total number of fragments amplified for all the 7 clones studied. Figure 2 showed the segregating markers among the sibs of each of the families amplified with primer OPE13.
|
|
|
Fig.2 RAPD profiles of five families amplified with primer OPE13 The arrows show the fragments that are polymorphic between parents and segregate among the sibs per family. In each family panel, M is the 100bp DNA ladder size standard(MBI)(order as Figure 1), P1 the female parent, P2 the male parent, and the rest 01~10 are sibs. |
Heterozygosity was defined as the percentage that heterozygous individuals took at a specific locus of the total individuals in a population (Nei, 1978). It was originally a concept of population genetics and later extended to refer the percentage of heterozygous loci at individual level (Lanham, 1996; Marshall et al., 1994). In our observations, high levels of heterozygosity were revealed in the parental eucalypt clones, with an average of 28. 0%(72 257). The heterozygosity varied with parent, as PB1(PC1, Clone 03) ranked the first in percentage of heterozygous loci (39.5%) but PD1(Clone 10) the last (16.2%). The levels of heterozygosity of eucalypts in this study were much higher than those estimated in nearly isogenic line of canola (Brassica napus)(3.6% at the most) (Marshall et al., 1994), and similar to those of Ribes nigrum(about 20%) (Lanham, 1996)
Interestingly, heterozygous loci or the number varied in the same parental clone with family (Table 4). For example, clone 03 showed 14 heterozygous loci in family B, but 9(one being exclusive) in family C. This might be caused by: (1) the disability of RAPD technology in detecting heterozygous loci in case of AA ×Aa, where the locus would not segregate in the sibs, and (2) the utilization of not-large-enough size of F1 population as some factual segregating loci could not be observed in a small population. The ideal solutions could rely on utilization of codominant markers, e. g. RFLP, and or selfed populaions in this term. However, dominant markers were employed on many occasions, and selfed crosses were difficult to be obtained due to self-incompatibility in most tree species (Marshall et al., 1994; Dudley et al., 1991). The usual populations used were single outcross pedigree in related studies. In addition, the probability for detecting a heterozygous loci in a population could be given by the probability of a fragment being present in at least one sib as 1-(1/2)n-1 when Aa ×aa (or aa ×Aa) and 1-(3/4)n-1 when Aa ×Aa, where n was the size of population. For a population of 10 sibs, the probability was 99.8% and 92.5% respectively. So a population size of 10 sibs could be effective for detecting possible segregating RAPD loci as was the case in this study.
In genetic mapping of highly heterozygous species, such as forest trees, the usual mapping population was F1 pedigree and the strategy was followed in a pseudo-testcross configuration. In such a strategy, the marker available for linkage map construction should be present in the mapping parent (genotype Aa)but absent in another (genotype aa) and segregate 1:1 in the sibs in a Mendelian fashion (Carlson et al., 1991; Grattapaglia et al., 1994). The high level of heterozygosity in eucalypt clones studied may indicate that testcross cases for RAPD markers were so common in eucalypt and the lack of codominance with RAPD might not represent such a disadvantage in genetic map construction. We could suggest that preliminary screening of small samples of progeny be conducted to determine the parental polymorphism and heterozygosity prior to fullscale segregating analysis. In this respect, Nps(Table 2) could be referable in choosing a rational F1 population and those fragments polymorphic between parents and segregant among sibs be potentially useful for genetic mapping in E. urophylla and E. tereticornis. Therefore, family E (Nps = 19), followed with families C and D (Nps=15), would be probably sound candidates of plant materials for genetic mapping (with an appropriate number of sibs, e. g. 100 or more). It might be inferred from this study that more than 300 primers could be selected out of 1 000 candidate primers and about 500 markers be produced for genetic mapping of a typical eucalypt species. If these primers were utilized in combination, even more markers could be mapped (Carlson et al., 1991).
In summary, the results from the present study indicate that high levels of polymorphism ad heterozygosity are reliably revealed in parental eucalypt clones with RAPD markers and thus facilitate the construction of genetic maps for plant material preparation considerations.
Acknowledgements
Thanks are due to (China) National Science Foundation Committee (grant No. 39870619)and International Foundation for Science (grant No. D 2947-1) for financial support in this study.
甘四明, 施季森, 白嘉雨, 等. 1998. 林木分子标记研究进展. 林业科学研究, 11: 428-434. DOI:10.3321/j.issn:1001-1498.1998.04.015 |
甘四明, 施季森, 白嘉雨. 2000. Idaho Rapidcycler PCR仪上用薄壁管进行RAPD扩增的程序优化. 南京林业大学学报, 24: 27-31. |
甘四明, 施季森, 白嘉雨, 等. 1999. 尾叶桉和细叶桉无性系的RAPD指纹图谱构建. 南京林业大学学报, 23: 11-14. |
徐建民, 白嘉雨, 甘四明. 1996. 尾叶桉家系综合选择的研究. 林业科学研究, 9: 561-567. DOI:10.3321/j.issn:1001-1498.1996.06.001 |
吴坤明, 吴菊英, 徐建民, 等. 1996. 桉树种间杂种优势和优良杂种的评选. 广东林业科技, 12: 1-5. |
Carlson J E, Tulsieram L K, Glaubitz J C, et al. 1991. Segregation of random amplified DNA markers in F1 progeny of conifers. Theor Appl Genet, 83: 194-200. DOI:10.1007/BF00226251 |
Doudrick R L, Nance W L, Nelson C D, et al. 1993. Detection of DNA polymorphisms in a single urediniospore-derived culture of Cronartium quercuum f. sp. fusiforne. Phytopathology, 83: 388-392. DOI:10.1094/Phyto-83-388 |
Doyle J J, Doyle J L. 1990. Isolation of plant DNA from fresh tissue. Focus, 12: 13-14. |
Dudley J W, Maroof M A S, Rufener G K. 1991. Molecular markers and grouping of parents in maize breeding programs. Crop Science, 31: 718-723. DOI:10.2135/cropsci1991.0011183X003100030036x |
Eldridge K, Davidson J, Harwood C, et al. 1993. Eucalypt Domestication and Breeding. Oxford: Oxford University Press.
|
Grattapaglia D and Sederoff R. 1994. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross mapping strategy and RAPD markers. Genetics, 137: 1121-1137. |
Hamrick J L and Godt M J W. Allozyme diversity in plant species. In Brown AHD, Clegg M T, and Kahler A L et al(eds.) Plant Population Genetics, Breeding and Genetic Resources. Sinaeur Association Massachusetts, 1990: 43-63
|
Jacobs M R. 1981. Eucalyptus Planting. Rome: FAO UN: 622-626. |
Lanham P G. Estimation of heterozygosity in Ribes nigrum L. using RAPD markers. Genetica. |
Lanza L L B, Souza Jr C L de, Ottoboni L M M, et al. 1997. Genetic distance of inbred lines and prediction of maize single-cross performance using Rapd markers. Theor Appl Genet, 94: 1023-1030. DOI:10.1007/s001220050510 |
Marshall P, Marchand M C, Lisieczko Z, et al. 1994. A simple method to estimate the percentage of hybridity in canola (Brassica napus) F1 hybrids. Theor Appl Genet, 89: 853-858. |
Martin G B, Brommonschenkel S H, Chungwondse J, et al. 1993. Map-base cloning of a protein kinase gene conferring disease resistance in tomato. Science, 262: 1432-1435. DOI:10.1126/science.7902614 |
Nei M. 1987. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89: 583-590. |
Welsh J, McClelland M. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucl. Acids Res., 18: 7213-7218. DOI:10.1093/nar/18.24.7213 |
Williams J G K, Kubelik A R, Livak K J, et al. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl. Acids Res., 18: 6531-6535. DOI:10.1093/nar/18.22.6531 |
 2003, Vol. 39
2003, Vol. 39