文章信息
- Tang Li, Tang Fang, Duan Jinghua, Liu Youquan, Zhong Qiuping
- 唐丽, 唐芳, 段经华, 刘友全, 钟秋平
- Cloning and Sequence Analysis of a Homologous Linalool Synthase Gene Involved in Floral Scents in Osmanthus fragrans var. thunbergii
- 金桂芳樟醇合成酶基因的克隆与序列分析
- Scientia Silvae Sinicae, 2009, 45(5): 11-19.
- 林业科学, 2009, 45(5): 11-19.
-
文章历史
- 收稿日期:2009-01-22
-
作者相关文章
2. 中国林业科学研究院林业研究所 北京 100091;
3. 中国林业科学研究院经济林研究开发中心 郑州 450003;
4. 中国林业科学研究院亚热带林业实验中心 分宜 336600
2. Research Institute of Forestry, CAF Beijing 100091;
3. Non-Timber Forestry Research and Development Center, CAF Zhengzhou 450003;
4. The Experimental Centre of Subtropical Forestry, CAF Fenyi 336600
Many plants had specific floral scent that is usually used for attracting specific pollinators. The floral scent compounds can also be used in perfumery, foods and medicines. Previous studies showed that most floral scent compounds belong to three major groups:phenylpropanoids, fatty acid derivatives and terpenoids (Croteau et al., 1991). Monoterpenoids are the simplest class of terpenoids as they contain only 10 carbon atoms. Most members of the monoterpenoids are constructed by the monoterpene synthases that catalyze the conversion of geranyl diphosphate, the universal, acyclic C10 intermediate of isoprenoid biosynthesis, to the cyclic parents of the various monoterpene skeletal types (Croteau et al., 1991). The crucial role of the cyclases in the origin of the different monoterpene structural groups has stimulated considerable interest in the details of these related enzymatic transformations as a model for the general terpenoid synthase reaction type (Raguso et al., 1996).
The activity of terpenoid synthase has been investigated from various plant species including Lavandula angustifolia, Clarkia breweri and Mentha citrate etc. And several full length cDNAs encoding monoterpene synthases involved in floral scents have been cloned and characterized (Christian et al., 2007; Wang et al., 1997; Crowell et al., 2002; Michal et al., 2002). Of the monoterpenoids, a major constituent is linalool. Linalool synthase gene (Lis), is targeted to plastids and catalyzes the conversion of geranyl diphosphate (GPP) to the monoterpene, linalool, in a single step (Dudareva et al., 1996). GPP, the precursor of monoterpenes, is in turn produced by GPP synthase from isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP) in both the plastids and cytosol of photosynthetic and non-photosynthetic cells (Phillips et al., 1999).
As one of the Chinese traditional famous flowers, Osmanthus fragrans has been in cultivation in China for over 2 000 years. Since ancient times, great attentions have been paid to the Osmanthus fragrans for its great economic, ornamental, medicinal and economic value. Osmanthus fragrans var. thunbergii, a gold-orange variety, emits a stronger floral scent than other varieties of the Osmanthus fragrans. Flowers of Osmanthus fragrans var. thunbergii are vivid yellowish orange in color and bloom from late September to mid-October in Hunan area.
The floral scent production of Osmanthus fragrans var. thunbergii has been investigated. And dozens of components were identified by gas chromatography-mass spectrometry (GC-MS) analysis. A major component of the floral scent is linalool, an acyclic monoterpene common to the floral scents of numerous other plant species (Li et al., 2008; Tian et al., 2008). However, the molecular mechanism of biosynthesis of the floral scent component is unclear yet. Therefore, the objective of this study iscloning and sequencing of the gene associated with floral scent compounds, accordingly providing a basis for regulation and control of floral scent compounds in fragrant plant varieties.
1 Materials and methods 1.1 Plant materialsExperiment materials consisted of flowers and leaves of Osmanthus fragrans var. thunbergii. Flowers were harvested in 2-day-old (initial-bloom stage), 5-day-old (mid-bloom stage) and 9-day-old (late-bloom stage), respectively. All of the plant materials were collected in test site of Hunan Academy of Forestry, Changsha, Hunan Province.
1.2 Methods 1.2.1 Preparation of total RNATotal RNA was isolated from leaves and floral tissues of Osmanthus fragrans var. thunbergii using Micro-to-Midi Total RNA Purification System (Invitrogen) according to the manufacture's instructions. The quality of RNA was determined spectrophotometricaly, and by agarose gel electrophoresis. Purity was estimated from the absorbency ratios A260/A230 and A260/A280, a measure of contamination by the polyphenols/carbohydrates and proteins, respectively.
1.2.2 RT-PCRTo clone partial monoterpene synthase sequences, RT-PCR was carried out. First-strand cDNA was synthesized by RT from total RNA using the First Strand cDNA Synthesis Kit (MBI). The RT was performed in a total volume of 20 μL containing 2 μL 10×RNA PCR Buffer, 4 μL MgCl2 (25 mmol·L-1), 2 μL dNTP Mixture (dATP, dCTP, dGTP and dTTP, each at 10 mmol·L-1), 1 μL M-MuLV Reverse Transcriptase (5 U·μL-1), 0.5 μL RNase Inhibitor (40 U·μL-1), 1 μL Oligo(dT)18 primer (2.5 μmol·L-1), 2 μL RNA (around 1 μg), and 7.5 μL RNase-free water. The analysis was programmed in a thermal cycler (PE-9600, USA) and conducted under condition of 10 min at 30 ℃, 25 min at 50℃ and 5 min at 95℃. The second-step PCR amplification was performed using two newly designed primer combinations OfLisPF1 [5′-GATT(A/T)CAACTACAT(C/T)CA(A/G)TC(C/T)CT-3′]/OfLisPR1(5′-ATATTGAA(G/A)TCGAGTTT(G/T)GC(G/A)AG-3′) and OfLisPF1/OfLisPR2(5′-AAATC(C/T)(A/G)CCCAC(G/T)ATCTCTG(T/G)AA-3′). The primers were designed based on multiple amino acid sequence alignment of monoterpene synthases from Antirrhinum majus (GenBank accession No.EF433762), Arabidopsis thaliana (AF497485), Artemisia annua(AF154124 and AF154125), Backhousia citriodora (AB438045), Clarkia breweri (U58314 and AF067603), Clarkia concinna (AF067602), Lavandula latifolia (DQ421801 and DQ263741), Mentha citrate (AY083653), Perilla citriodora (AY917193), Picea abies (AY473623) and Perilla frutescens var. crispa (AF444798).
For expression detection of the putative linalool synthase gene in different tissues of Osmanthus fragrans var. thunbergii, RT-PCR was conducted with specific primer combination OfLisPF2 (5′-GATGATAACTACATCCAATCTCT-3′)/OfLisPR3 (5′-CTTGCTAAACTCGATTTCAATAT-3′), which spanned the region of 659 bp in the linalool synthase gene.
The PCR cycling parameters consisted of 38 cycles of denaturation for 30 s at 94 ℃, annealing for 30 s at 58 ℃, and extension for 5 min at 72 ℃.
1.2.3 RACE strategyTo clone the full length cDNA of the putative terpene synthase gene, RACE strategy was adopted based on known partial sequences acquired by RT-PCR. The 3′-ends of the gene were amplified with 3′RACE System for Rapid Amplification of cDNA Ends (Invitrogen, Version E). Total RNA was reverse transcribed using SuperScriptTM Ⅱ Reverse Transcriptase with Adapter Primer provided in the system under condition of 10 min at 70℃, 50 min at 42℃, 15 min at 70℃ and 20 min at 37 ℃. The second PCR amplification was conducted with primers OfLisGSP3F and AUAP. OfLisGSP3F (5′-TTGGAGGATTCAGAGGCTAGAG-3′) was designed on the basis of the partial sequences for the putative OfLis gene, and AUAP was provided in the 3′RACE System. The amplification program consisted of 35 cycles of 30 s at 94℃, 30 s at 59℃, 3 min at 72℃, and a final extension of 7 min at 72℃.
The 5′-ends of the gene were amplified with 5′RACE System for Rapid Amplification of cDNA Ends (Invitrogen, Version 2.0). First-strand cDNA was synthesized by RT using SuperScriptTM Ⅱ with primer OfLisGSP5R1 (5′-CTTGCTAAACTCGATTTCAATAT-3′). Then dc-tailed cDNA was amplified by using the Abridged Anchor Primer (AAP, 5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGG IIG-3′, provide in the System) and nested OfLisGSP5R2 (5′-AAGCTCAAGGATAACTTG GTTCAT-3′). Finally, the cDNA product was reamplified using Abridged Universal Amplification Primer (AUAP, 5′-GGCCACGCGTCGACTA GTAC-3′, provide in the System) and nested primer OfLisGSP5R3 (5′-GTGCCTATCTTGTTTTTCATACATA-3′). The latter two PCR reactions were programmed as 3′ RACE-PCR.
1.2.4 Cloning, sequencing and sequence analysisRT-PCR and RACE products were analyzed by 1.5% agarose gel. Subsequently, the target products were extracted and purified from 2% agarose gel using a Gel Extraction Kit (Ambiogen, China), TA-ligated into pMD18-T Vector (Takara, Japan), and transformed into Escherichia coli strain DH5α. The positive clones were screened by the white/blue colony, and identified by plasmid PCR with consensus primers M13PM4 (forward, 5′-GTTTTCCCAGTCACGAC-3′) and M13PRV (5′-CAGGAAACAGCTATGAC-3′). Target clones were sequenced in both directions by Shanghai Sangon Biological Engineering Technology & Services Co., Ltd.
A preliminary analysis of DNA sequence data was performed with Vector NTI. The BLAST algorithm was used to search the NCBI GenBank (http://www.ncbi.nlm.nih.gov/) databases for homologous sequences. Prediction of isoeletric point, molecular weight and Prosite was performed by using online ExPASy proteomics tools (http://expasy.org/tools/pi_tool.html). Motif analysis of the target gene was conducted by Motif Scan (http://www.myhits.isb-sib.ch/) and sequence comparison with related genes. Signal peptide was predicted by Signal 3.0 Server(http://www.cds.dtu.dk/services/SignalP). Hydrophobicity analysis was carried out by online ProScale tool (http://www.expasy.org/cgi-bin/protscale.pl). Multiple amino acid sequence alignment was performed with the Clustal W method by Align X program using software DNAMAN. Phylogenetic tree was constructed by neighbour-joining method using Vector NTI.
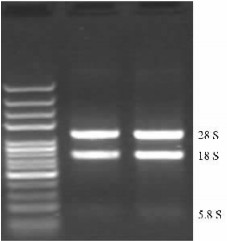
2 Results 2.1 Isolation of linalool synthase gene fragment in Osmanthus fragrans var. thunbergiiUsing Total RNA Purification System (Invitrogen), the total RNA was successfully isolated from flowers of Osmanthus fragrans var. thunbergii. The RNA sample had a ratio OD 260/280 of 1.905 6, suggesting the RNA was not contaminated by DNA. The RNA quality was verified by visualization on the agarose gel (Fig. 1), which showed no degradation and was of high quality allowing us to use it in the next RT-PCR amplification.
|
Fig.1 Electrophoresis detection of total RNA from Osmanthus fragrans var. thunbergii flowers |
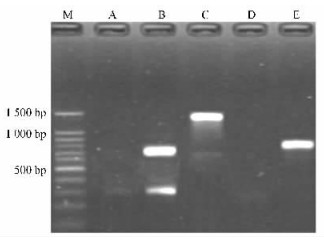
Following RT-PCR with two newly designed primer combinations, the combination of OfLisPF1/OfLisPR2 was found to be most successful, amplifying two fragments that included a strong one of approximately 700 bp and a weak one of around 300 bp (Fig. 2). Taking the region between OfLisPF1 and OfLisPR2 into consideration, only the 700 bp-sized fragment seemed to represent linalool synthase gene. After cloning and sequencing the two PCR products, two distinct nucleotide sequences were determined. When using the nucleotide sequence for the about 300 bp-sized fragment as the query by Blastn, it was shown that no significant similar sequence was found in GenBank database. In contrast, a number of monoterpene synthase sequences were searched out in a Blast search of GenBank using the nucleotide sequence of the about 700 bp-sized fragment as the query. In light of the denomination convention of published linalool synthase genes in other plant species, the 700 bp-sized gene fragment was designated as OfLis (Osmanthus fragrans var. thunbergii linalool synthase gene) and submitted to GenBank database (GenBank accession No. FJ645727).
|
Fig.2 cDNA amplification of OfLis gene M:100 bp DNA ladder; A:RT-PCR with OfLisPF1/OfLisPR1;B:RT-PCR with OfLisPF1/OfLisPR2;C:3′RACE-PCR with OfLisGSP3F/AUAP; D:5′RACE-PCR with AAP/OfLisGSP5R2;E:5′RACE-PCR with AUAP/OfLisGSP5R3. |
Following 3′ RACE-PCR, an approximately 1 300 bp-sized fragment was produced (Fig. 2). Considering the location of the OfLisGSP3F primer in linalool synthase gene, the 1 300 bp amplification product should represent the 3′-end fragment of the OfLis. In 5′ RACE-PCR, no visible amplification fragment was observed in agarose gel when using AAP/OfLisGSP5R2 primer combination. Using the PCR product amplified by primer combination AAP/OfLisGSP5R2 as template DNA, nested PCR was conducted with AUAP/OfLisGSP5R3. Consequently, an approximately 800 bp fragment was produced. Sequencing result showed that the 5′ and 3′ RACE-PCR amplification fragments were 798 bp and 1 269 bp, respectively, and both of them corresponded to the OfLis gene by Blastn analysis. The two nucleotide sequences shared the common region of 64 bp, and the OfLis full length cDNA was determined by overlapping the sequences of 5′ and 3′ ends accordingly.
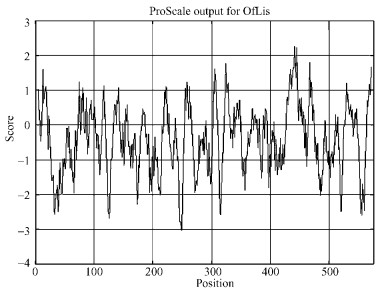
2.3 Sequence characterization of OfLisThe OfLis full length cDNA was 2 003 bp in length and contained a complete open reading frame (ORF) of 1 731 bp encoding 576 amino acids (Fig. 3). It contained a 48 bp 5′-untranslated region (5′ UTR). The putative initiation codon ATG was at positions 49-51. The termination codon TGA was present at positions 1777-1779, followed by 3′ UTR, which contained the consensus polyadenylation signal AATAAA at positions 1833-1838. The tail-adding signal is likely to help mRNA get across nucleus, and plays an important role in steadying mRNA and improving efficiency of translation. The OfLis presented two typical conserved motifs of terpene synthases, i.e DDXXD and (N, D)D(L, I, V)X(S, T)XXXE, which are involved in the coordination of divalent cations and thereby are essential for substrate binding and ionization. It also displayed the N-terminal peptide sequence RR(X)8W that is essential for the enzymatic activity of many monoterpene synthases. LQLYEASYLL is another typical motif frequently found in monoterpene synthases, but slightly altered in OfLis gene where the amino acid residue Leu was replaced by Val. Amino acid sequence comparison of OfLis with other monoterpene synthases showed that the biggest differences between monoterpene synthases were located upstream of RR(X)8W motif (Fig. 4). OfLis contained 35 amino acids in which comparatively many serine and alanine residues and few acidic amino acids were integrated. The characters were often present in signal peptides, which likely directed proteins to plastids where they were processed to their active mature forms by truncation of the N-terminal peptides. OfLis contained such a plastidial signal peptide, confirming that it belongs to monoterpene synthases. Signal 3.0 Server also predicted a signal peptide of 23 amino acids, which is consistent with type and number of amino acid residue upstream of RR(X)8W motif. The molecular weight and isoeletric point of OfLis were predicted to be 67.29 ku and 5.26, respectively. Analyzed by ProScale, it was shown that most of the amino acid sequences of OfLis were hydrophilic regions (Fig. 5).

|
Fig.3 Nucleotide and amino acid sequences of OfLis cDNA The predicted amino acid sequence is shown directly below the nucleotide sequence. The asterisk indicates the termination codon. The consensus polyadenylation signal AATAAA is underlined. Four characteristic motifs are shaded. |

|
Fig.4 Amino acid sequence comparison of OfLis with other linalool genes Four characteristic motifs are boxed. Aa:Artemisia annua; Bc:Backhousia citriodora; La:Lavandula angustifolia; Ll:Lavandula latifolia; Mc:Mentha citrate; Pa:Picea abies; Pc:Perilla citriodora; Pf:Perilla frutescens var. crispa. |
|
Fig.5 Hydrophobicity plot analysis of the predicted polypeptide of OfLis The abscissa denotes the position of amino acid residues in the OfLis. The ordinate denotes the hydrophobic and hydrophilic score. The hydrophobic and hydrophilic positions are plotted above and below the ordinate, respectively. |
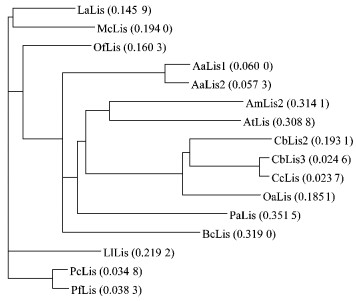
A total of 16 amino acid sequences for monoterpene synthases were aligned by clustalx. Similarity analysis indicated that OfLis exhibited the highest similarity (63.6%) with Lavandula angustifolia linalool synthase, and the lowest (19.0%) with Clarkia concinna linalool synthase gene. Based on the multiple amino acid sequence alignment, a neighbour-joining phylogenetic tree was generated. In accord with similarity scores, OfLis displayed the closest relatedness with Lavandula angustifolia linalool synthase on the phylogenetic tree (Fig. 6).
|
Fig.6 Phylogenetic tree depicting relatedness of linalool synthase genes Aa:Artemisia annua; Am:Antirrhinum majus; At:Arabidopsis thaliana; Bc:Backhousia citriodora; Cb:Clarkia breweri; Cc:Clarkia concinna; La:Lavandula angustifolia; Ll:Lavandula latifolia; Mc:Mentha citrate; Oa:Oenothera arizonica; Pa:Picea abies; Pc:Perilla citriodora; Pf:Perilla frutescens var. crispa. |
To detect tissue expression specificity of OfLis, total RNA was isolated from Osmanthus fragrans var. thunbergii leaves and floral tissues (sepals, petals, pistils and stamens) of 2-day-old (initial-bloom stage), 5-day-old (mid-bloom stage) and 9-day-old (late-bloom stage) flowers. RT-PCR revealed that a 700 bp fragment with expected size was generated from the total RNA of petals, pistils and stamens, whereas no amplification product was observed in leaves and sepals. Subsequently, the 700 bp-sized fragment was cloned and sequenced. Sequence analysis revealed that it corresponded to Oflis gene, suggesting that the Oflis gene was specifically in petals, pistils and stamens. In addition, the 700 bp fragment was detected in three stages, indicating that Oflis was expressed in the whole bloom stage.

|
Fig.7 RT-PCR for detection of tissue expression of OfLis Ⅰ:Floral tissues of 2-day-old; Ⅱ:Floral tissues of 5-day-old; Ⅲ: Floral tissues of 9-day-old. M:100 bp DNA ladder; A:Leaves; B:Sepals; C:Petals; D:Pistils; E:Stamens. |
Previous studies have shown that volatile compounds associated with floral scent can be grouped into more than a dozen chemical classes, including organic acids, aldehydes, ketones, alcohols, esters, lactones, sulfur compounds, acetals, furans, phenols, terpenes, and epoxides. Early research on floral scent initially focused on identification of flavor components and subsequently on characterizing the volatiles that convey the characteristic odor unique to a particular flower and unraveling their biogenesis.
To clarify the mechanism of biosynthesis, regulation and control of floral scent components biosynthesis on molecular level, researchers have cloned a few genes directly influencing floral scent. The first terpene synthase genes were cloned from tobacco and spearmint (Mentha spicata)(Facchini et al., 1992; Colby et al., 1993). According to their sequence information, related genes were isolated and cloned from other species, e.g. from Arabidopsis thaliana (Bohlmann et al., 2000; Chen et al., 2003; 2004;Faldt et al., 2003; Tholl et al., 2005), Clarkia breweri (Dudareva et al., 1996), Abies grandis (Bohlmann et al., 1997; 1998; 1999), Ocimum basilicum (Iijima et al., 2004), Salvia officinalis (Wise et al., 1998), Mentha spicata (Colby et al., 1993), Citrus limon (Lucker et al., 2002) and Zea mays (Schnee et al., 2002; Kollner et al., 2004).
In this study, we successfully cloned a linalool synthase gene, OfLis, by degenerate primer design and RACE strategy from Osmanthus fragrans var. thunbergii flowers. The OfLis cDNA appeared to encode a typical N-terminal plastidial targeting sequence, which was anticipated for a monoterpene synthase. And the expected DDXXD motif was present in the C-terminal domain which played an essential role in substrate binding and ionization. The cDNA also encoded tandem arginine residues near the N terminus, which were highly conserved in the monoterpene cyclases and were necessary for the conduct of the initial isomerization of geranyl diphosphate that must precede the normally coupled cyclization catalyzed by this enzyme type. Although function of the OfLis gene in biosynthesis of floral scent compounds has not been investigated, these sequence characters strongly support that the OfLis encodes typical linalool synthase and is likely to play a significant role in regulation and control of floral scent compounds in Osmanthus fragrans var. thunbergii flowers.
To our knowledge, this is the first report of gene associated with floral scent in Osmanthus fragrans. The OfLis sequences cloned in this study lay the basis for elucidating the synthesis mechanism of monoterpenoids in Osmanthus fragrans var. thunbergii flowers and for genetic modification of flavor profile of this economically important tree species.
Bohlmann J, Steele C L, Croteau R, et al. 1997. Monoterpene synthases from grand fir (Abies grandis) cDNA isolation, characterization, and functional expression of myrcene synthase, (-)-(4S)-limonene synthase, and (-)-(1S, 5S)-pinene synthase. Chem, 272: 21784-21792. |
Bohlmann J, Meyer-Gauen G, Croteau R. 1998. Plant terpenoid synthases:Molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA, 95: 4126-4133. DOI:10.1073/pnas.95.8.4126 |
Bohlmann J, Phillips M, Ramachandiran V, et al. 1999. cDNA cloning, characterization, and functional expression of four new monoterpene synthase members of the Tpsd gene family from grand fir (Abies grandis). Arch Biochem Biophys, 368: 232-243. DOI:10.1006/abbi.1999.1332 |
Bohlmann J, Martin D, Oldham N J, et al. 2000. Terpenoid secondary metabolism in Arabidopsis thaliana cDNA cloning, characterization, and functional expression of a myrcene/(E)-beta-ocimene synthase. Arch Biochem Biophys, 375: 261-269. DOI:10.1006/abbi.1999.1669 |
Chen F, Ro D K, Petri J, et al. 2004. Characterization of a root-specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1, 8-cineole. Plant Physiol, 135: 1956-1966. DOI:10.1104/pp.104.044388 |
Chen F, Tholl D, D'Auria J C, et al. 2003. Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell, 15: 481-494. DOI:10.1105/tpc.007989 |
Christian L, Barbara F, Maria F, et al. 2007. Cloning and functional characterization of three terpene synthases from lavender (Lavandula angustifolia). Archives of Biochemistry and Biophysics, 465: 417-429. DOI:10.1016/j.abb.2007.06.011 |
Colby S M, Alonso W R, Katahira E J, et al. 1993. 4S-limonene synthase from the oil glands of spearmint (Mentha spicata). cDNAisolation, characterization, and bacterial expression of the catalytically active monoterpene cyclase. Biol Chem, 268: 23016-23024. |
Croteau R, Karp F. 1991. Origin of natural odorants//Muller P, Lamparsky D. Perfume: Art, Science and Technology. Elsevier Applied Sciences, New York, 101-126.
|
Crowell A L, Williams D C, Davis E M, et al. 2002. Molecular cloning and characterization of a new linalool synthase. Archives of Biochemistry and Biophysics, 405: 112-121. DOI:10.1016/S0003-9861(02)00348-X |
Dudareva N, Cseke L, Blanc V M, et al. 1996. Evolution of floral scent in clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell, 8: 1137-1148. DOI:10.1105/tpc.8.7.1137 |
Facchini P J, Chappell J. 1992. Gene family for an elicitor-induced sesquiterpene cyclase in tobacco. Proc Natl Acad Sci, 89: 11088-11092. DOI:10.1073/pnas.89.22.11088 |
Faldt J, Arimura G I, Gershenzon J, et al. 2003. Functional identification of AtTPS03 as (E)-β- ocimene synthase: a monoterpene synthase catalyzing jasmonate- and wound-induced volatile formation in Arabidopsis thaliana. Planta J, 216: 745-751. |
Iijima Y, Davidovich-Rikanati R, Fridman E, et al. 2004. Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol, 136: 3724-3736. DOI:10.1104/pp.104.051318 |
Kollner T G, Schnee C, Gershenzon J, et al. 2004. The variability of sesquiterpenes emitted from two Zea mays cultivars is controlled by allelic variation of two terpene synthase genes encoding stereoselective multiple roduct enzymes. Plant Cell, 16: 1115-1131. DOI:10.1105/tpc.019877 |
Li Zuguang(李祖光), Cao Hui(曹慧), Zhu Guohua(朱国华), et al. 2008. Study on chemical constituents of fragrance released from fresh flowers of three different Osmanthus fragrans Lour. during different florescences. Chemistry and Industry of Forestry Products(林产化学与工业), 28(3): 75-80. http://en.cnki.com.cn/Article_en/CJFDTOTAL-LCHX200803018.htm
|
Lucker J, Tamer M K, Schwab W, et al. 2002. cDNA isolation and functional analysis of four monoterpene synthases. Biochem, 269: 3160-3171. |
Michal L, Amir Z, Efraim L, et al. 2002. Linalool and linalool oxide production in transgenic carnation flowers expressing the Clarkia breweri linalool synthase gene. Molecular Breeding, 9: 103-111. DOI:10.1023/A:1026755414773 |
Phillips M A, Croteau R. 1999. Resin-based defenses in conifers. Trends Plant Sci, 4: 184-190. DOI:10.1016/S1360-1385(99)01401-6 |
Raguso R A, Light D M. 1996. Electroantennogram responses of Hyles lineata (Sphingidae:Lepidoptera) to volatile compounds from Clarkia breweri (Onagraceae) and other moth-pollinated flowers. J Chem Ecol, 22: 1735-1766. DOI:10.1007/BF02028502 |
Schnee C, Kollner T G, Gershenzon J, et al. 2002. The maize gene terpene synthasel encodes a sesquiterpene synthase catalyzing the formation of (E)-farnesene, (E)-nerolidol, and (E, E)-farnesol after herbivore damage. Plant Physiol, 130: 2049-2060. DOI:10.1104/pp.008326 |
Tian Guanghui (田光辉), Liu Cunfang (刘存芳), Gu Tianqi (辜天琪), et al. 2008. Analysis of aroma components from the senescence flowers of Osmanthus fragrans by GC-MS. Journal of Anhui Agri Sci (安徽农业科学), 36(17): 7214-7216.
|
Tholl D, Chen F, Petri J, et al. 2005. Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J, 42: 757-771. DOI:10.1111/tpj.2005.42.issue-5 |
Wang Jihong, Natalia D, Shyam B, et al. 1997. Floral scent production in Clarkia breweri (Onagraceae) Ⅱ. Localization and developmental modulation of the enzyme S-adenosyl-L-methionine: (iso)eugenol O-methyltransferase and phenylpropanoid emission. Plant Physiology, 24: 418-526. |
Wise M L, Savage T J, Katahira E, et al. 1998. Monoterpene synthases from common sage (Salvia officinalis) cDNA isolation, characterization, and functional expression of (+)-sabinene synthase, 1, 8-cineole synthase, and (+)-bornyl diphosphate synthase. Chem, 273: 14891-14899. |
 2009, Vol. 45
2009, Vol. 45
