文章信息
- Zhang Lin, Tan Xiaofeng, Zhou Jian, He Xiaoyong, Yuan Deyi, Hu Jiao, Long Hongxu
- 张琳, 谭晓风, 周建, 何小勇, 袁德义, 胡姣, 龙洪旭
- Determination of S-Genotypes of Seven Cultivars and Identification of a Novel S-RNase Allele in Pyrus pyrifolia
- 7个砂梨品种S基因型的确定及1个新S-RNase基因的分离鉴定
- Scientia Silvae Sinicae, 2008, 44(10): 42-48.
- 林业科学, 2008, 44(10): 42-48.
-
文章历史
- 收稿日期:2008-02-25
-
作者相关文章
2. 河南科技学院园林学院 新乡453003;
3. 浙江省丽水市科普工作指导站 丽水 323000
2. Botanical Garden Institute, Henan Institute of Science and Technology Xinxiang 453003;
3. Lishui Popular Science Station of Zhejiang Province Lishui 323000
Self-incompatibility (SI) is a genetically controlled mechanism which is adopted by a large number of flowering plants to prevent inbreeding and promote out-crossing (de Nettancourt, 1977). The SI in Rosaceae, Solanaceae and Scrophurariaceae families belongs to the type of gametophytic SI (GSI) that is controlled by the single multi-allelic S-locus containing two separate genes, i.e. one stylar S-gene and one pollen S-gene (Anderson et al., 1986; Sassa et al., 1992; Xue et al., 1996).
Japanese pear (Pyrus pyrifolia) was the first species of the Rosaceae family in which stylar S-locus allele specific proteins were identified as S-RNases with ribonuclease activity. To date, a total of 10 S-RNases were identified in Japanese pear and their primary structure was determined (Ishimizu et al., 1998a; 1998b; Castillo et al., 2002; Takasaki et al., 2004). Knowledge of S-genotype is apparently essential for pear production and breeding programs. The conventional pollination-based methods for cultivars S-genotyping are time consuming and labor intensive. Moreover, the obtained results are often ambiguous due to environmental and physiological factors. PCR-based molecular techniques provide a rapid, reliable and easily manipulated approach for cultivars S-genotyping of pear including Japanese pear, Korean-bred pear (P. pyrifolia) and European pear (P. communis).
Chinese sand pear (P. pyrifolia) is one of the major dominant pear species in China with a great number of excellent cultivars and genetic resources (Luo et al., 2002).It mainly distributes in South of China and has been considered to be the same species as similar germplasm in Japan (Teng et al., 2002). In recent years, many Chinese-bred pear cultivars using cultivars from Chinese sand pear or Japanese pear as the parents have been released which inherit good features from sand pear. Previous researches on SI of sand pear in China allowed determination of a few cultivars and isolation of dozens of putative new S-RNase alleles (Tan et al., 2005; Wuyun et al., 2007). In the present study, we selected seven cultivars that are important in pear production or pear breeding to analyze their S-genotypes, aiming to facilitate the pear production and breeding.
1 Materials and methods 1.1 Plant materialsSeven pear cultivars`Hangqing', `Cuiguan', `Xizilv', `Chubixiang', `Huangpishui', `Zhenghedaxueli' and`Hongtaiyang' were used in this study. Young leaves of these cultivars were collected in spring and stored at-80 ℃. Flowers of 'Zhenghedaxueli' were collected at balloon stage. Styles and anthers were removed and immediately frozen in liquid nitrogen and stored at-80 ℃. All plant materials were provided by Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences (CAAS).
1.2 Isolation of nucleic acidsGenomic DNAs were isolated from young leaves of the seven cultivars with a CTAB method as described by Tan et al. (2007). Total RNAs were isolated from leaves, anthers and styles of`Zhenghedaxueli' using Micro-to-Midi Total RNA Purification System (Invitrogen) and following the manufacture's protocols. The quality of DNAs and RNAs was checked by electrophoresis in 0. 8% TAE agarose gel.
1.3 Amplification of S-RNase alleles from genomic DNAsPCR was conducted from genomic DNAs of the seven cultivars with PF1 and PR1. PF1 (5′-TTTACGCAGCAATATCAG-3′, forward primer) and PR1 (5′-AC(A/G)TT(C/T)GGCCAAATA(A/G)TT-3′, reverse primer) spanned HV region in the sequence of pear S-RNases and were designed based on the conserved amino acid sequences`FTQQYQ' from conserved region 1 (C1), and`anti-(I/T)IWPNV' between the C2 and C3 conserved regions downstream of the intron insertion point. The PCR reaction was programmed as described by Tan et al. (2007). The amplification products were detected by 1.8% agarose gel electrophoresis. Separated bands produced in each reaction were extracted and purified using a Gel Extraction Kit (Ambiogen, China). The products assayed as a single band in agarose gel were further separated by 6% non-denaturing PAGE followed by ethidium bromide (EB) staining (Chen, 2006). The separated bands were extracted with the method of Chen (2006). As the yield of the extracted DNA from polyacrylamide gel was low, this DNA was used as a template for a second PCR with PF1 and PR1. To extend the sequence of newly identified S43-RNase allele, primer pair PF2 and PR1 was employed for genomic PCR. PF2 (5′-TGCCTCGCTCTTGAACAAA-3′) was designed based on the conserved nucleotide sequences between the initiation codon (ATG) and the putative TATA box in the 5′ flanking region (Castillo et al., 2002). The amplification conditions are the same as above. The target amplification fragment was extracted and purified from 1.8% agarose gel as described above.
1.4 RT-PCRReverse transcription (RT) was performed using a cDNA amplification Kit (Takara, Japan) according to the manufacturer's directions from total RNAs from leaves, anthers and styles, respectively. The reaction was programmed for 1 cycle of 30 ℃ 10 min, 50 ℃ 25 min, and 95 ℃ 5 min. The second-step PCR amplification was carried out with primer combination PF2 and PR2 for 38 cycles of denaturation for 30 s at 94 ℃, annealing for 30 s at 55 ℃, and extension for 2 min at 72 ℃. PR2 (5′-GTGATTTTATCGTCTGAGCAT-3′), a S43-specific reverse primer, was designed for distinguishing S43 from S13 based on nucleotide sequence alignment.
1.5 Cloning, sequencing and sequence analysis of PCR productsThe purified PCR products were TA-ligated into pMD18-T Vector (Takara, Japan) and transformed into Escherichia coli strain DH5α. The positive clones are screened by the white/blue colony, and identified by plasmid PCR with BcaBESTTM Sequencing Primer M13-47 (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) and BcaBESTTM Sequencing Primer RV-M (5′-GAGCGGATAACAATTTCACACAGG-3′). Target clones were sequenced in both directions by Shanghai Biosune Biotechnology Ltd.
Analysis of the DNA sequences determined was conducted with BLAST software (http://www.ncbi.nlm.nih.gov/BLAST/). The deduced amino acid sequences were aligned with the method of ClustalW by software Vector NTI. Similarities between S-RNase alleles were computed by Vector NTI and BLASTP. Prediction of signal peptide and prosite was conducted by using online software ExpASy (http://expasy.org/).
2 Results 2.1 Length polymorphism of amplification fragments in different cultivarsAfter PCR amplification with consensus primers PF1 and PR1 for pear S-RNase gene, the amplified products were analyzed by 1.8% agarose gel electrophoresis. Fig. 1 showed that two differently sized bands ranging from approximately 360 bp to 1 000 bp were generated in each of the four cultivars `Chubixiang', `Huangpishui', `Zhenghedaxueli' and`Hongtaiyang'. However, the other three cultivars showed a distinct banding pattern, i. e. only a single band of around 370 bp was observed in each cultivar. Subsequently, the PCR products of the three cultivars were further electrophoresed on 6% non-denatured polyacrylamide gel. Consequently, two fragments were successfully separated in each of the three cultivars (Fig. 2). The six DNA fragments extracted from polyacrylamide gel were used as templates for a second PCR with PF1 and PR1 to increase DNA yield. Finally, a total of 14 amplification fragments were successfully cloned and sequenced.

|
Fig.1 Analysis of PCR products of seven pear cultivars by 1.8% agarose gel electrophoresis M: 100 bp DNA ladder (Oumay, China); A:'Hangqing'; B:'Cuiguan'; C:'Xizilv'; D:'Chubixiang'; E:'Huangpishui'; F:'Zhenghedaxueli'; G:'Hongtaiyang'. The PCR products were amplified with PF1 and PR1. |

|
Fig.2 Analysis of PCR products of three pear cultivarsby 6% PAGE M: 100 bp DNA ladder (Oumay, China); A:'Hangqing'; B:'Cuiguan'; C:'Xizilv'. The PCR products were amplified with PF1 and PR1. |
Comparison of nucleotide sequences revealed that two or more than two sequences were found to be identical among the 14 sequences which were classified into 10 different types accordingly. Although the region between PF1 and PR1 does not cover a complete gene, BLASTn analysis revealed that all of the 10 sequences displayed the typical structural organization of pear S-RNase with conserved and HV regions, of which nine correspond to known pear S-RNase alleles S1, S4, S5, S8, S13, S15, S16, S35 and S42, respectively. The other one sequence with a unique size (494 bp) identified in'Zhenghedaxueli', however, showed significant differences to all published pear S-RNases, thus it was considered to be a new S-RNase allele sharing the highest similarity (88%) with P. pyrifolia S8-RNase at the nucleotide level. This new allele was temporarily named as S43-RNase (GenBank accession No.EF566873) following S42-RNase in GenBank database.
Based on the sequence analysis of S-RNase alleles cloned here, the S-genotypes of the seven cultivars were unambiguously determined as follows:'Hangqing' (S1S4), 'Cuiguan' (S1S5), 'Xizilv' (S1S4), 'Chubixiang' (S1S15), 'Huangpishui' (S16S42), 'Zhenghedaxueli' (S13S43) and'Hongtaiyang' (S8S35).
2.3 Organ expression detection and sequence characterization of the new S43-RNaseTo extend the sequence of the new S43 allele, PF2 were employed for PCR amplification. Following analysis by 1.8% agarose gel electrophoresis, two bands including a strong one of around 600 bp and a weak one of approximately 480 bp were observed (Fig. 3). As is shown in Fig. 1, S43 is longer than S13, thus the 600 bp-sized fragment should represent the S43 and the 480 bp one appearing as a faint band should be S13. After cloning the target fragment of 600 bp, three independent clones were sequenced. Sequencing result showed that the three clones were identical, indeed corresponding to S43.
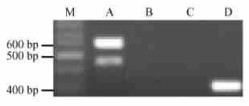
|
Fig.3 Genomic and cDNA sequences amplification of S43-RNase M: 100 bp DNA ladder (Oumay, China); A: Genomic sequence extension of S43-RNase with PF2 and PR1; B-D: RT-PCR on total RNAs from leaves (B), anthers (C) and styles (D) with PF2 and PR2. |
To clone the cDNA sequence of S43 and investigate its organ expression character, RT-PCR was carried out from the total RNAs from leaves, anthers and styles of'Zhenghedaxuele', respectively. Consequently, only one product of approximately 300 bp was amplified from stylar RNA, but not from leaves and anthers (Fig. 3). After the cDNA fragment was cloned and sequenced, the sequence was found to well match the S43-RNase. Comparison of cDNA and genomic sequences of S43-RNase revealed that it contained an intron of 294 bp.
The deduced amino acid sequence of S43-RNase was established and then aligned other pear S-RNases and its sequence was characterized as shown in Fig. 4. In comparison with known pear S-RNases, S43-RNase displayed the typical structural features of pear S-RNases, i.e. a signal peptide, two conserved regions (C1 and C2) and one HV region. S43-RNase showed one histidine residue (His-60) essential for T2/S type RNase activity (Kawata et al., 1990) and presented three cysteine residues mostly conserved in S-RNases. The partial amino acid sequence for S43-RNase also showed three potential N-glycosilation sites whose glycans might be important in the folding and stabilization of the core structure (Ishimizu et al., 1999). Alignment of the deduced amino acid sequence of the S43-RNase with published Maloideae S-RNases sequence showed the highest similarity (92%) with sand pear S8-RNase, and the lowest similarity (65%) with wild apple (Malus transitoria) St-RNase (GenBank accession No. AB035928).
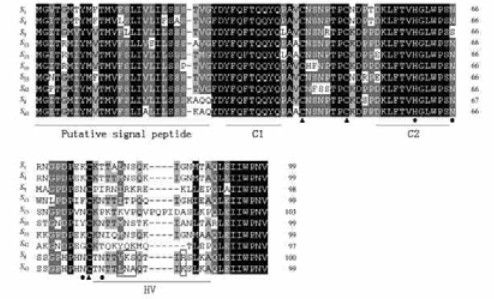
|
Fig.4 Alignment of the deduced amino acid sequences of the cloned 10 S-alleles and characterization of the new S43-RNase The amino acid residues conserved among the 10 S-alleles are shaded. The putative signal peptide, a hypervariable (HV) region and two conserved regions (C1 and C2) are underlined. For S43-RNase, conserved cysteine residues, histidine residues essential for the RNase activity and potential N-glycosylation sites are marked with symbols ▲, ◆ and ●, respectively. |
The consensus primers PF1 and PR1 have been proved to be suitable for S-RNase allele amplification from Japanese pear cultivars. The results presented herein together with the published data (Tan et al., 2005; Wuyun et al., 2007) suggest that they can also be employed for S-RNase allele identification from Chinese sand pear and interspecific hybrid. And the partial fragments for S-RNase alleles amplified from genomic DNAs can be distinguished by agarose gel electrophoresis or PAGE based on the intron length variation.
During the preparation of the present manuscript, the three cultivars 'Hangqing', 'Xizilv' and'Cuiguan' were S-genotyped with the method of endonuclease digestion of PCR products. Gu (2006) genotyped the three cultivars as S3S7, S1S4 and S1S4S5, respectively, while Wuyun et al. (2007) genotyped the latter two cultivars as S1S4 and S3S5, respectively.`Hangqing' is a natural hybrid of'Chili', and it was considered to be the pollen parent of'Yaqing' ('Yali''Hangqing') and'Xinhang' ('Shinseiki''Hangqing'). The S-genotype of`Chili' has not been reported yet but, Heng et al. (2007) assigned'Yaqing' as S4S17 and inferred that'Yaqing' should inherit the S4 from'Hangqing'. Moreover, the genotype of S1S3 was assigned to'Xinhang' also by Gu (2006). The S3 allele should be from'Shinseiki' because 'Shinseiki' has been genotyped as S3S4 in Japan (Hiratsuka et al., 1998). The other one allele S1, obviously, should be given by'Hangqing'. From the analysis above, therefore, our assignment of'Hangqing' as S1S4 seems to be correct. The S-genotype of'Xizilv' identified here is the same with that reported by Gu (2006) and Wuyun et al. (2007) but, a surprise comes from the pedigree analysis. The'Xizilv' is considered to be the cross'Shinseiki' ('Yakumo' 'Hangqing') and the S-genotype of'Yakumo' was S1S4 (Castillo et al., 2001), implying that'Yakumo' is compatible with'Hangqing' although they shared an identical S-genotype. This finding would be useful in pear breeding program.'Cuiguan' is the progeny of'Kosui' (S4S5) (Castillo et al., 2001) ('Hangqing''Shinseiki'). This cultivar was S-genotyped as S1S4S5, and identified as a diploid but not a triploid by flow cytometry by Gu (2006). Analysis of the pedigree of`Cuiguan' showed that all of the three S-genotypes of S1S4S5, S3S5 and the S1S5 appear to be reasonable, thus the discrepancy might be due to mislabeling samples in the orchard.'Cuiguan' is an important cultivar with high-quality grown in South of China. Therefore, further work will be required for correct assignment of S-genotype to'Cuiguan' in which additional methods can be adopted such as pollination tests.'Hongtaiyang' is a new and quality pear cultivar from interspecific hybridization of'Meigetsu' (P. pyrifolia)'Flemish beauty' (P. communis) which was bred in China. The S-genotypes of'Meigetsu' and'Flemish beauty' were reported to be S1S8 (Castillo et al., 2001) and SdSe (Takasaki et al., 2006). Comparative analysis show that there are only three nucleotide differences between S35 and Sd out of the 897 nucleotides compared, and the two alleles have an identical amino acid sequence (Fig. 5). Thus S35 is the same allele with Sd, although they are identified from different pear species by different researchers. From the S-genotype information of'Hongtaiyang', we can conclude that S8 is inherited from'Meigetsu' and the S35 (Sd) from'Flemish beauty', further confirming that the S-allele is indeed inherited in a Mendelian manner (Ishimizu et al., 1999).

|
Fig.5 Sequence comparison of sand pear S35 (GenBank accession No.DQ839240) and European pear Sd (GenBank accession No.AB236427) |
A and B represents sequence comparison at the nucleotide and the deduced amino acid level, respectively. The differences are boxed. The identical nucleotides or amino acid residues were shadowed between the two S-RNase genes.
In this study, a new S43 allele was identified in Chinese sand pear. After the detection of gene organ expression by RT-PCR together with sequence analysis, S43 was proved to be an S-RNase gene which is specifically expressed in the style. It is suggested that two amino acid variations in HV region are sufficient for distinguishing different alleles in pear (Ishimizu et al., 1998b). S43 shows the highest similarity with pear S8 and four amino acid variations were present in their HV regions (Fig. 4), which is the evidence that S43 is considered to be a new S-allele. The new S43-RNase allele has hitherto been identified in the single cultivar'Zhenghedaxueli', implying that this cultivar should be compatible with all of the pear cultivars with known S-genotypes and is a promising cultivar used as a good pollination tree in pear production and breeding programs.
Anderson M A, Cornish E C, Mau S L, et al. 1986. Cloning of cDNA for a stylar glycoprotein associated with expression of self-incomp atibility in Nicotiana alata. Nature, 321: 38-44. DOI:10.1038/321038a0 |
Castillo C, Takasaki T, Saito T, et al. 2001. Reconsi deration of S-genotype assignments and discovery of a new allele based on S-RNase PCR-RFLPs in Japanese pear cultivars. Breeding Sci, 51: 5-11. DOI:10.1270/jsbbs.51.5 |
Castillo C, Takasaki T, Saito T, et al. 2002. Cloning of the S8-RNase (S8-allele) of Japanese pear (Pyrus pyrifolia Nakai). Plant Biotechnol, 19(1): 1-6. DOI:10.5511/plantbiotechnology.19.1 |
Chen Yonghua (陈永华). 2006. Studies on the physiological and molecular mechanism of submergence tolerance in rice (Oryza sativa L. ). Changsha: Hunan Agricultural University (Doctoral dissertation).
|
de Nettancourt D. 1977. Incompatibility in Angiosperms. Berlin: Springer-Verlag.
|
Gu Qingqing (辜青青). 2006. Determination of self-incompatibility genotypes in some Japanese pears (Pyrus pyrifolia Nakai) by PCR-RFL P. Wuhan: Huazhong Agricultural University (Master thesis).
|
Heng Wei (衡伟), Zhang Shaoling (张绍铃), Zhang Yuyan (张妤艳), et al. 2007. Identification of S-genotypes of twelve pear cultivars by analysis of DNA sequence. Acta Hortic Sinica (园艺学报), 34(4): 853-858. http://en.cnki.com.cn/Article_en/CJFDTotal-YYXB200704009.htm
|
Hiratsuka S, Kubo T, Okada Y. 1998. Estimation of self-incompatibility genotype in Japanese pear cultivars by stylar protein analysis. J Japan Soc Hort Sci, 67(4): 491-496. DOI:10.2503/jjshs.67.491 |
Ishimizu T, Shinkawa T, Sakiyama F, et al. 1998a. Primary structural features of rosaceous S-RNases associated with gametophytic self-incompatibility. Plant Mol Biol, 37: 931-941. DOI:10.1023/A:1006078500664 |
Ishimizu T, Endo T, Yamaguchi-Kabata Y, et al. 1998b. Identification of regions in which positive selection may operate in S-RNase of Rosaceae: implication for S-allele-specific recognition sites in S-RNase. FEBS Lett, 440: 337-342. DOI:10.1016/S0014-5793(98)01470-7 |
Ishimizu T, Inoue K, Shimonaka M, et al. 1999. PCR-based method for identifying the S-genotypes of Japanese pear cultivars. Theor Appl Genet, 98: 961-967. DOI:10.1007/s001220051156 |
Kawata Y, Sakiyama F, Hayashi F, et al. 1990. Identification of two essential histidine residues of ribonuclease T2 from Aspergillus oryzae. Eur J Biochem, 187: 255-262. DOI:10.1111/ejb.1990.187.issue-1 |
Luo Z R, Zhang Q L. 2002. The genetic resources and their utilization of Pyrus pyrifolia in China. Acta Hort, 634: 201-205. |
Sassa H, Hirano H, Ikehashi H. 1992. Self-incompatibility-related RNase in style of Japanese pear (Pyrus serotina Rehd.). Plant Cell Physiol, 33: 811-814. |
Takasaki T, Moriya Y, Okada K, et al. 2006. cDNA cloning of nine S alleles and establishment of a PCR-RFLP system for genotyping European pear cultivars. Theor Appl Genet, 112: 1543-1552. DOI:10.1007/s00122-006-0257-7 |
Takasaki T, Okada K, Castillo C, et al. 2004. Sequence of the S9-RNase cDNA and PCR-RFLP system for discriminating S1- to S9-allele in Japanese pear. Euphytica, 135: 157-167. DOI:10.1023/B:EUPH.0000014907.50575.d0 |
Tan Xiaofeng (谭晓风), Wuyun Tana (乌云塔娜), Nakanishi T, et al. 2005. Identification and sequencing of seven new S-RNase genes from Pyrus pyrifolia. J Cent South Forest Univ (中南林学院学报), 25(4): 1-6. http://en.cnki.com.cn/Article_en/CJFDTotal-ZNLB200504000.htm
|
Tan Xiaofeng, Zhang Lin, Wuyun Tana, et al. 2007. Molecular identification of two new self-incompatible alleles (S-alleles) in Chinese pear (Pyrus bretschneideri). J Plant Physiol Mol Biol, 33: 61-70. |
Teng Y, Tanabe K. 2002. Reconsideration on the origin of cultivated pears native to east Asia. Acta Hort, 634: 151-155. |
Wuyun Tana (乌云塔娜), Tan Xiaofeng (谭晓风), Li Xiugen (李秀根), et al. 2007. Identification of S-genotypes of 13 progenies of 'Shinseiki' pear. Scientia Silvae Sinicae (林业科学), 43(9): 116-122. http://www.linyekexue.net/CN/Y2007/V43/I09/116
|
Xue Y B, Carpenter R, Dickinson H G, et al. 1996. Origin of allelic diversity in Antirrhinum S-locus RNases. Plant Cell, 8: 805-814. DOI:10.1105/tpc.8.5.805 |
 2008, Vol. 44
2008, Vol. 44

