2. 海军军医大学(第二军医大学)第二附属医院骨肿瘤科, 上海 200003;
3. 海军军医大学(第二军医大学)第一附属医院泌尿外科, 上海 200433
2. Department of Orthopaedic Oncology, The Second Affiliated Hospital of Naval Medical University (Second Military Medical University), Shanghai 200003, China;
3. Department of Urology, The First Affiliated Hospital of Naval Medical University (Second Military Medical University), Shanghai 200433, China
尤文肉瘤(Ewing’s sarcoma,EWS)是发生于儿童和青少年的第二常见恶性原发性骨肿瘤,占所有儿科恶性肿瘤的3%[1]。过去几十年中EWS的治疗取得较大进展,除手术、放射治疗、化学治疗等常规治疗手段外,以靶向和免疫治疗为主的综合治疗方式在改善EWS患者预后方面成效显著,使早期患者5年总生存率达到70%,但由于EWS高度异质性和侵袭性,导致约80%的患者在诊断初期即发生局部进展或远处转移事件,该类患者的5年总生存率通常低于30%[2]。因此,亟须探究更为精准的诊断方法及治疗靶点。
染色体易位是EWS的基因组特征之一,其中t(11;22)(q24:q12)易位占比最大,该易位将组成型表达的尤文肉瘤断裂区域1(Ewing sarcoma breakpoint region 1,EWSR1)的氨基末端反式激活结构域与很少表达的佛氏白血病病毒整合蛋白1(Friend leukemia virus integration 1,FLI1)的羧基末端DNA结合结构域融合在一起,形成EWSR1-FLI1融合蛋白[3]。EWSR1-FLI1融合蛋白是EWS的关键促癌分子,具有多重功能和复杂的调控机制。代谢重编程是指在病理状态下,细胞通过多种调控机制改变其代谢途径和产物分布,以适应其生长和生存需要的过程。这种代谢途径的变化不仅可以影响细胞内营养物质的利用,还可以影响其周围微环境的营养和代谢物质的流通[4]。代谢重编程被认为是肿瘤发生和发展的关键过程[5]。随着代谢组学与高通量测序技术的发展,EWS相关代谢酶、代谢途径、代谢产物等方面的研究不断深入,学者们对EWS的发病机制有了新的认识[6]。本文对EWSR1-FLI1融合蛋白通过经典代谢方式影响EWS发生、发展的相关研究进展进行梳理和汇总,旨在深入探讨EWS的代谢重编程特征及潜在机制,为了解该病的发病机制、寻找新的诊断和治疗方法提供有用信息。
1 糖代谢葡萄糖代谢是维持细胞生长的基本代谢途径之一,包含多种合成和分解途径。糖的合成代谢主要包括糖异生和糖原合成;而分解代谢主要包括细胞质中的糖酵解途径、磷酸戊糖途径(pentose phosphate pathway,PPP)、丝氨酸合成途径,以及线粒体中的三羧酸循环(tricarboxylic acid cycle,TCA循环)和氧化磷酸化过程。其中,糖酵解作为核心代谢途径,其中间产物还可作为分支代谢途径的起点,如葡萄糖-6-磷酸可进入PPP,而3-磷酸甘油酸则可进入丝氨酸合成途径[7]。
正常细胞增殖程度有限,而肿瘤细胞增殖迅速且不受限制,因此肿瘤细胞必须产生更多能量以满足其增殖需要。无论氧气含量是否充足,肿瘤细胞总是倾向于通过无氧糖酵解而非有氧氧化产能(Warburg效应)[8]获得能量。目前已经确认Myc、Ras等多种癌基因可通过缺氧诱导因子(hypoxia inducible factor,HIF)影响肿瘤的糖代谢水平。在缺氧条件下,HIF-1α降解减少,葡萄糖转运体1(glucose transporter 1,GLUT1)表达水平显著提高,使肿瘤细胞能够更迅速地摄取葡萄糖[9]。Ceranski等[10]对156例EWS患者队列的基因表达水平与生存率进行量化,发现HIF-1α和GLUT1高表达与患者预后呈负相关。除GLUT1和HIF-1α等经典的糖代谢相关分子以外,EWSR1-FLI1融合蛋白也是调控EWS糖代谢重编程的关键分子。Tanner等[11]发现,EWSR1-FLI1基因敲除细胞与对照细胞的代谢谱有明显差异,线粒体应激试验表明EWSR1-FLI1基因敲除增加了呼吸和糖酵解功能;其中磷酸甘油酸脱氢酶(phosphoglycerate dehydrogenase,PHGDH)在EWS中高表达,并与患者生存率降低相关。EWSR1-FLI1融合蛋白可通过多种形式激活糖酵解途径(图 1):(1)EWSR1-FLI1融合蛋白可调控Ras-MAPK和PI3K/Akt信号通路,上调HIF-1α等转录因子的表达,促进HIF-1转录调控复合物形成及胞内积累,进而激活包括GLUT1在内的糖酵解相关酶的表达,增加肿瘤细胞对葡萄糖的摄取和代谢[12-13];(2)EWSR1-FLI1可与转录因子特异性蛋白1发生互作,或拮抗let-7,解除Ras对GLUT1的负向调控作用,最终上调GLUT1的表达水平[14];(3)EWSR1-FLI1还可以调控GLUT1的亚细胞定位和活性,影响肿瘤细胞对葡萄糖的摄取和利用[15]。在糖异生和PPP中,EWSR1-FLI1可以抑制糖异生途径中关键基因磷酸烯醇丙酮酸羧激酶和葡萄糖-6-磷酸酶的表达减少糖异生,上调血小板型磷酸果糖激酶和肝脏磷酸果糖激酶等糖酵解途径关键酶的表达,促进肿瘤细胞对葡萄糖的摄取和利用,并提高糖酵解途径中的产物输出[16-17];下调葡萄糖-6-磷酸脱氢酶[18]和6-磷酸葡萄糖酸内酯酶[19]等关键酶的表达,导致PPP通路下调,使肿瘤细胞更依赖糖酵解途径,进而获取大量ATP和还原型烟酰胺腺嘌呤二核苷酸磷酸。
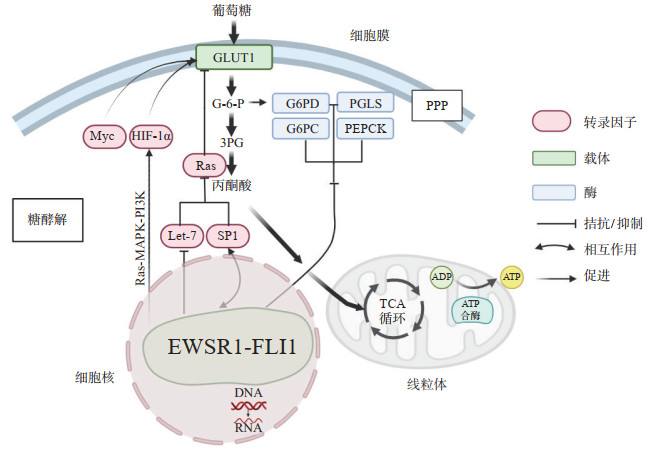
|
图 1 EWSR1-FLI1融合蛋白影响糖代谢的途径 EWSR1:尤文肉瘤断裂区域1;FLI1:佛氏白血病病毒整合蛋白1;PPP:磷酸戊糖途径;GLUT1:葡萄糖转运体1;HIF-1α:缺氧诱导因子1α;MAPK:丝裂原活化蛋白激酶;PI3K:磷脂酰肌醇3-激酶;SP1:特异性蛋白1;3PG:3-磷酸甘油酸;G-6-P:葡萄糖-6-磷酸;G6PD:葡萄糖-6-磷酸脱氢酶;PGLS:6-磷酸葡萄糖酸内酯酶;G6PC:葡萄糖-6-磷酸酶;PEPCK:磷酸烯醇丙酮酸羧激酶;TCA循环:三羧酸循环;ADP:腺苷二磷酸;ATP:腺苷三磷酸. |
基于EWS相关糖代谢重编程特征,已有一些特异性抑制剂和治疗方法得以研究和应用。常见的糖酵解抑制剂包括2-脱氧-D-葡萄糖(2-deoxy-D-glucose,2-DG)、3-溴丙酮酸等。2-DG能与GLUT1结合,经己糖激酶磷酸化为2-DG-6-磷酸,使GLUT1不能进一步代谢,从而阻断糖酵解途径。Dasgupta等[20]的研究数据显示,用2-DG预处理3 d能够使EWS细胞生长显著延迟(P<0.000 1)。HIF-1α通过调节多种代谢途径、生物过程和细胞功能促进EWS的发展,因此被视为EWS的重要治疗靶点。目前靶向HIF-1α的药物正在陆续研发中,但仍处于早期开发阶段,深入研究HIF在肿瘤代谢中的作用将为进一步开发HIF抑制剂提供新的思路[10, 21]。靶向GLUT的药物也是抗肿瘤药物研究的热点,如BAY-876、WZB117、STF-31等[22]。Ojelabi等[23]研究显示,WZB117通过抑制人红细胞和癌细胞系中的被动糖转运,并限制糖酵解抑制小鼠肿瘤生长;Kraus等[24]通过短时[18F]-氟代脱氧葡萄糖摄取实验发现,在50 μmol/L浓度下,糖酵解抑制剂STF-31能够显著抑制肿瘤细胞的葡萄糖摄取。目前关于这些抑制剂对EWS的抗肿瘤作用的报道不多,仍有待进一步研究和探索。
2 氨基酸代谢重编程氨基酸代谢可以提供能量和碳源,肿瘤细胞在缺乏营养和氧气的情况下更倾向于利用氨基酸进行能量代谢[7]。在EWS中,EWSR1-FLI1融合蛋白可调节氨基酸合成相关酶的表达,如上调谷氨酰胺酶(glutaminase,GLS)[25]、磷酸丝氨酸转氨酶1(phosphoserine aminotransferase 1,PSAT1)[26]、天冬酰胺合成酶(asparagine synthetase,ASNS)[27]等,下调谷氨酸脱氢酶(glutamate dehydrogenase,GDH)[28]、PHGDH[11]等,从而使氨基酸合成和积累增多,促进肿瘤增殖和生长。Sen等[26]研究发现EWSR1-FLI1可正向调节丝氨酸-甘氨酸生物合成所需蛋白质的表达和替代营养源谷氨酰胺的摄取,EWS细胞的存活依赖于谷氨酰胺,而EWSR1-FLI1可上调谷氨酰胺转运蛋白[钠-葡萄糖协同转运蛋白2(sodium-glucose transporter 2,SGLT2;也称溶质载体家族(solute carrier family,SLC)5A2]和参与单碳循环的两种酶[亚甲基四氢叶酸脱氢酶1(methylenetetrahydrofolate dehydrogenase 1,MTHFD1)、亚甲基四氢叶酸脱氢酶1样蛋白(methylenetetrahydrofolate dehydrogenase 1-like,MTHFD1L)]的表达;抑制EWS细胞中的丝氨酸-甘氨酸生物合成会影响其氧化还原状态,导致活性氧积累、DNA损伤和细胞凋亡。EWSR1-FLI1融合蛋白还可促进氨基酸转运体SLC7A5和SLC1A5的表达,增加EWS细胞对氨基酸的摄取[29]。
氨基酸分解代谢是一种通过转氨酶作用将氨基酸中的氨基与α-酮酸结合,生成新的氨基酸和新的酮酸的代谢途径。在EWS细胞中,EWSR1-FLI1融合蛋白主要通过抑制酮体途径中的丙酮酸脱氢酶[30]和乳酸脱氢酶[31]的表达,降低氨基酸转化为丙酮酸的速率,从而促进肿瘤的生长和侵袭。此外,Mutz等[32]研究发现,在EWSR1-FLI1融合蛋白低表达的EWS细胞A673sh中,色氨酸-2, 3-双氧合酶激活,导致色氨酸降解,犬尿氨酸和犬尿喹啉酸水平升高,犬尿氨酸/犬尿喹啉酸依赖的芳香烃受体信号激活,使A673sh细胞在EWSR1-FLI1融合蛋白低表达条件下存活,这可能代表肿瘤细胞存活的一种新的替代途径。
针对EWS的氨基酸代谢,目前的治疗方法主要集中在两个方面:抑制氨基酸合成和促进氨基酸分解。Olszewski等[33]研究表明,使用二氯乙酸抑制丙酮酸脱氢酶激酶处理后,EWS细胞的增殖和侵袭行为被有效抑制。Issaq等[34]发现,PHGDH抑制剂NCT-503能够剂量依赖性地抑制EWS细胞增殖;烟酰胺磷酸核糖转移酶(nicotinamide phosphoribosyltransferase,NAMPT)抑制剂GNE-618能够阻断PHGDH底物烟酰胺腺嘌呤二核苷酸的产生,将GNE-618与NCT-503联合使用可发挥协同作用,消除EWS细胞增殖和肿瘤生长。此外,一些靶向氨基酸代谢相关酶的药物,如哺乳动物雷帕霉素靶蛋白复合物1/2(mammalian target of rapamycin complex 1/2,mTORC1/2)抑制剂、PI3K/Akt通路抑制剂、氨基酸转运蛋白抑制剂等,也被作为EWS的潜在抗肿瘤药物受到广泛关注[35-36]。Eyre等[37]进行的Ⅱ期临床试验观察了口服vistusertib(mTORC1/2双联抑制剂)对复发或难治性弥漫大B细胞淋巴瘤的疗效,28例患者接受了vistusertib治疗,最长周期仅为19 d,参与治疗的患者已有部分缓解。Banerji等[38]的临床研究表明,在给予AZD5363(一种选择性泛Akt抑制剂)一段时间后,实体肿瘤患者群体(n=90)的肿瘤缩小,并且有27例(30%)和6例(7%)患者分别在≥6周和≥12周内病情稳定。虽然目前的研究结果显示通过抑制氨基酸合成、促进氨基酸分解可以有效抑制EWS细胞的生长和侵袭,但尚未见能够应用于临床的有效药物,仍需进一步研究。
3 脂代谢和核苷酸代谢重编程除了糖代谢和氨基酸代谢外,脂代谢和核苷酸代谢也是肿瘤细胞代谢的重要组成部分。脂代谢包括脂肪酸的合成和分解。EWS细胞的脂代谢变化可促进细胞增殖和转移,并参与药物抵抗。在脂肪酸合成途径中,EWSR1-FLI1融合蛋白可促进脂肪酸合成酶(fatty acid synthase,FASN)的表达[39]。此外,哺乳动物雷帕霉素靶蛋白信号通路的激活也可促进脂肪酸合成酶的表达及活性,并抑制脂肪酸的氧化代谢和分解,最终促进脂肪酸的合成和存储[40]。在脂肪酸分解中,由于EWS细胞受丙酮酸含量影响,影响丙酮酸合成与转化的生物学过程都将影响脂解代谢。丙酮酸通过酯化反应转化为甘油三酯,进而生产维持EWS细胞生长所必需的脂质成分[41]。
针对EWS的脂代谢重编程,FASN抑制剂如奥美拉唑、TVB-2640(地尼凡他)等,因其可限制肿瘤细胞获取足够的脂肪酸,进而减缓细胞的生长和增殖,目前已在临床前研究中取得一定疗效。Sardesai等[42]在三阴性乳腺癌患者中进行的Ⅱ期临床试验结果显示,奥美拉唑单药治疗组患者FASN表达显著降低(P=0.02),且奥美拉唑耐受性良好,并无≥3级的不良反应。Kelly等[43]开展的Ⅱ期临床试验对25例复发性高级别星形细胞瘤患者进行TVB-2640联合贝伐珠单抗治疗,结果显示患者总缓解率达56%。
核苷酸代谢包括核苷酸的合成和分解途径,在维持细胞增殖和生存过程中发挥重要作用。肿瘤细胞的许多侵袭行为,包括不受控制的增殖、化疗抵抗、免疫逃逸和转移等,都与增强的核苷酸代谢有关[44]。对肿瘤中核苷酸代谢的认识揭示了其对免疫逃逸的非增殖性作用,提示抗核苷酸代谢药在增强免疫治疗中的潜在作用[45]。EWSR1-FLI1融合蛋白可能参与核苷酸合成的调控,如通过调节腺苷酸合成酶、鸟苷酸合成酶等核苷酸代谢相关酶的表达,从而促进核苷酸的合成[11, 46]。通过抑制核苷酸水解酶的活性可抑制肿瘤细胞的增殖和转移[47]。核苷类似物6-巯基嘌呤(6-mercaptopurine,6-MP)能够通过对代谢酶的竞争性抑制作用干扰或抑制嘌呤核苷酸的合成,因而具有抗肿瘤作用,一项Ⅱ期临床试验结果显示6-MP对EWS的真实缓解率不超过26%[48]。
4 小结代谢重编程在肿瘤发展、转移与耐药中的作用已得到越来越多的揭示,增进对肿瘤代谢重编程的理解能够为肿瘤的治疗提供更多策略。随着EWS中代谢相关机制研究的不断深入,最新的治疗方法从不同代谢途径入手,为EWS的免疫和靶向治疗提供了新的靶点,并开发出一些新的药物。但目前的研究多数仍处于体外研究水平,新药研发尚缺乏大量高级别有效证据,临床疗效证据不足,且部分受限于其安全性,一些已在其他肿瘤中应用的靶点抑制剂是否能够应用于EWS仍需进一步研究。EWS细胞代谢的复杂性决定了单一治疗方法可能无法彻底杀伤肿瘤,因此联合治疗可能更具前景,如联合应用氨基酸代谢抑制剂和糖代谢抑制剂,或化学治疗、放射治疗与氨基酸代谢抑制剂联合使用等,这还有待进一步研究。期望未来能有更多的研究揭示EWS代谢相关的潜在诊断和治疗靶点,研发出更有效的药物和治疗方案,从而提高EWS患者的生存率和生活质量。
| [1] |
JAIN S, KAPOOR G. Chemotherapy in Ewing's sarcoma[J]. Indian J Orthop, 2010, 44(4): 369-377. DOI:10.4103/0019-5413.69305 |
| [2] |
RIGGI N, SUVÀ M L, STAMENKOVIC I. Ewing's sarcoma[J]. N Engl J Med, 2021, 384(2): 154-164. DOI:10.1056/nejmra2028910 |
| [3] |
GORTHI A, ROMERO J C, LORANC E, et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma[J]. Nature, 2018, 555(7696): 387-391. DOI:10.1038/nature25748 |
| [4] |
OHSHIMA K, MORⅡ E. Metabolic reprogramming of cancer cells during tumor progression and metastasis[J]. Metabolites, 2021, 11(1): 28. DOI:10.3390/metabo11010028 |
| [5] |
VANDER HEIDEN M G, DEBERARDINIS R J. Understanding the intersections between metabolism and cancer biology[J]. Cell, 2017, 168(4): 657-669. DOI:10.1016/j.cell.2016.12.039 |
| [6] |
HONG B, LI Y, YANG R, et al. Single-cell transcriptional profiling reveals heterogeneity and developmental trajectories of Ewing sarcoma[J]. J Cancer Res Clin Oncol, 2022, 148(12): 3267-3280. DOI:10.1007/s00432-022-04073-3 |
| [7] |
LI Z, ZHANG H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression[J]. Cell Mol Life Sci, 2016, 73(2): 377-392. DOI:10.1007/s00018-015-2070-4 |
| [8] |
WARBURG O. On respiratory impairment in cancer cells[J]. Science, 1956, 124(3215): 269-270. DOI:10.1126/science.124.3215.269 |
| [9] |
BOSE S, LE A. Glucose metabolism in cancer[M]//LE A. The heterogeneity of cancer metabolism. Cham: Springer International Publishing, 2018: 3-12. DOI: 10.1007/978-3-319-77736-8_1.
|
| [10] |
CERANSKI A K, CARREÑO-GONZALEZ M J, EHLERS A C, et al. Hypoxia and HIFs in Ewing sarcoma: new perspectives on a multi-facetted relationship[J]. Mol Cancer, 2023, 22(1): 49. DOI:10.1186/s12943-023-01750-w |
| [11] |
TANNER J M, BENSARD C, WEI P, et al. EWS/FLI is a master regulator of metabolic reprogramming in Ewing sarcoma[J]. Mol Cancer Res, 2017, 15(11): 1517-1530. DOI:10.1158/1541-7786.MCR-17-0182 |
| [12] |
KILIC-EREN M, BOYLU T, TABOR V. Targeting PI3K/Akt represses hypoxia inducible factor-1α activation and sensitizes rhabdomyosarcoma and Ewing's sarcoma cells for apoptosis[J]. Cancer Cell Int, 2013, 13(1): 36. DOI:10.1186/1475-2867-13-36 |
| [13] |
LEE O W, RODRIGUES C, LIN S H, et al. Targeted long-read sequencing of the Ewing sarcoma 6p25.1 susceptibility locus identifies germline-somatic interactions with EWSR1-FLI1 binding[J]. Am J Hum Genet, 2023, 110(3): 427-441. DOI:10.1016/j.ajhg.2023.01.017 |
| [14] |
HAMEIRI-GROSSMAN M, PORAT-KLEIN A, YANIV I, et al. The association between let-7, RAS and HIF-1α in Ewing sarcoma tumor growth[J]. Oncotarget, 2015, 6(32): 33834-33848. DOI:10.18632/oncotarget.5616 |
| [15] |
ARYEE D N T, NIEDAN S, KAUER M, et al. Hypoxia modulates EWS-FLI1 transcriptional signature and enhances the malignant properties of Ewing's sarcoma cells in vitro[J]. Cancer Res, 2010, 70(10): 4015-4023. DOI:10.1158/0008-5472.CAN-09-4333 |
| [16] |
XING Y Q, LI A, YANG Y, et al. The regulation of FOXO1 and its role in disease progression[J]. Life Sci, 2018, 193: 124-131. DOI:10.1016/j.lfs.2017.11.030 |
| [17] |
HU-LIESKOVAN S, ZHANG J, WU L, et al. EWS-FLI1 fusion protein up-regulates critical genes in neural crest development and is responsible for the observed phenotype of Ewing's family of tumors[J]. Cancer Res, 2005, 65(11): 4633-4644. DOI:10.1158/0008-5472.CAN-04-2857 |
| [18] |
RORIE C J, THOMAS V D, CHEN P, et al. The Ews/Fli-1 fusion gene switches the differentiation program of neuroblastomas to Ewing sarcoma/peripheral primitive neuroectodermal tumors[J]. Cancer Res, 2004, 64(4): 1266-1277. DOI:10.1158/0008-5472.can-03-3274 |
| [19] |
GROHAR P J, GRIFFIN L B, YEUNG C, et al. Ecteinascidin 743 interferes with the activity of EWS-FLI1 in Ewing sarcoma cells[J]. Neoplasia, 2011, 13(2): 145-153. DOI:10.1593/neo.101202 |
| [20] |
DASGUPTA A, TRUCCO M, RAINUSSO N, et al. Metabolic modulation of Ewing sarcoma cells inhibits tumor growth and stem cell properties[J]. Oncotarget, 2017, 8(44): 77292-77308. DOI:10.18632/oncotarget.20467 |
| [21] |
BAN H S, UTO Y, WON M, et al. Hypoxia-inducible factor (HIF) inhibitors: a patent survey (2011-2015)[J]. Expert Opin Ther Pat, 2016, 26(3): 309-322. DOI:10.1517/13543776.2016.1146252 |
| [22] |
RECKZEH E S, WALDMANN H. Development of glucose transporter (GLUT) inhibitors[J]. Eur J Org Chem, 2020, 2020(16): 2321-2329. DOI:10.1002/ejoc.201901353 |
| [23] |
OJELABI O A, LLOYD K P, SIMON A H, et al. WZB117 (2-fluoro-6-(m-hydroxybenzoyloxy) phenyl m-hydroxybenzoate) inhibits GLUT1-mediated sugar transport by binding reversibly at the exofacial sugar binding site[J]. J Biol Chem, 2016, 291(52): 26762-26772. DOI:10.1074/jbc.M116.759175 |
| [24] |
KRAUS D, RECKENBEIL J, VEIT N, et al. Targeting glucose transport and the NAD pathway in tumor cells with STF-31: a re-evaluation[J]. Cell Oncol (Dordr), 2018, 41(5): 485-494. DOI:10.1007/s13402-018-0385-5 |
| [25] |
ZHANG H F, HUGHES C S, DELAIDELLI A, et al. Abstract PR002: identification of metabolic adaptation mechanisms that drive anoikis suppression and metastasis in Ewing sarcoma[J]. Cancer Res, 2023, 83(2 Suppl): PR002. DOI:10.1158/1538-7445.metastasis22-pr002 |
| [26] |
SEN N, CROSS A M, LORENZI P L, et al. EWS-FLI1 reprograms the metabolism of Ewing sarcoma cells via positive regulation of glutamine import and serine-glycine biosynthesis[J]. Mol Carcinog, 2018, 57(10): 1342-1357. DOI:10.1002/mc.22849 |
| [27] |
BELL R S, WUNDER J, ANDRULIS I. Molecular alterations in bone and soft-tissue sarcoma[J]. Can J Surg, 1999, 42(4): 259-266. |
| [28] |
BELYANSKAYA L. Expression and subcellular localization of Ewing sarcoma (EWS) protein is affected by the methylation process[J]. Exp Cell Res, 2003, 288(2): 374-381. DOI:10.1016/s0014-4827(03)00221-0 |
| [29] |
RATHORE R, CALDWELL K E, SCHUTT C, et al. Metabolic compensation activates pro-survival mTORC1 signaling upon 3-phosphoglycerate dehydrogenase inhibition in osteosarcoma[J]. Cell Rep, 2021, 34(4): 108678. DOI:10.1016/j.celrep.2020.108678 |
| [30] |
CERNIGLIA G, DEY S, GALLAGHER-COLOMBO S M, et al. Abstract B05: PI3K/mTOR pathway-dependent regulation of oxygen metabolism via pyruvate dehydrogenase (PDH)-E1alpha phosphorylation[J]. Mol Cancer Ther, 2015, 14(7 Suppl): B05. DOI:10.1158/1538-8514.pi3k14-b05 |
| [31] |
FLORES G, GROHAR P J. One oncogene, several vulnerabilities: EWS/FLI targeted therapies for Ewing sarcoma[J]. J Bone Oncol, 2021, 31: 100404. DOI:10.1016/j.jbo.2021.100404 |
| [32] |
MUTZ C N, SCHWENTNER R, KAUER M O, et al. EWS-FLI1 impairs aryl hydrocarbon receptor activation by blocking tryptophan breakdown via the kynurenine pathway[J]. FEBS Lett, 2016, 590(14): 2063-2075. DOI:10.1002/1873-3468.12243 |
| [33] |
OLSZEWSKI U, POULSEN T T, ULSPERGER E, et al. In vitro cytotoxicity of combinations of dichloroacetate with anticancer platinum compounds[J]. Clin Pharmacol, 2010, 2: 177-183. DOI:10.2147/CPAA.S11795 |
| [34] |
ISSAQ S H, MENDOZA A, KIDNER R, et al. EWS-FLI1-regulated serine synthesis and exogenous serine are necessary for Ewing sarcoma cellular proliferation and tumor growth[J]. Mol Cancer Ther, 2020, 19(7): 1520-1529. DOI:10.1158/1535-7163.MCT-19-0748 |
| [35] |
ZHANG S, LI D, JIAO G J, et al. miR-185 suppresses progression of Ewing's sarcoma via inhibiting the PI3K/AKT and Wnt/β-catenin pathways[J]. Onco Targets Ther, 2018, 11: 7967-7977. DOI:10.2147/OTT.S167771 |
| [36] |
ERKIZAN H V, KONG Y, MERCHANT M, et al. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing's sarcoma[J]. Nat Med, 2009, 15(7): 750-756. DOI:10.1038/nm.1983 |
| [37] |
EYRE T A, HILDYARD C, HAMBLIN A, et al. A phase Ⅱ study to assess the safety and efficacy of the dual mTORC1/2 inhibitor vistusertib in relapsed, refractory DLBCL[J]. Hematol Oncol, 2019, 37(4): 352-359. DOI:10.1002/hon.2662 |
| [38] |
BANERJI U, DEAN E J, PÉREZ-FIDALGO J A, et al. A phase Ⅰ open-label study to identify a dosing regimen of the pan-AKT inhibitor AZD5363 for evaluation in solid tumors and in PIK3CA-mutated breast and gynecologic cancers[J]. Clin Cancer Res, 2018, 24(9): 2050-2059. DOI:10.1158/1078-0432.CCR-17-2260 |
| [39] |
SHI X, ZHENG Y, JIANG L, et al. EWS-FLI1 regulates and cooperates with core regulatory circuitry in Ewing sarcoma[J]. Nucleic Acids Res, 2020, 48(20): 11434-11451. DOI:10.1093/nar/gkaa901 |
| [40] |
SUBBIAH V, BROWN R E, JIANG Y, et al. Morphoproteomic profiling of the mammalian target of rapamycin (mTOR) signaling pathway in desmoplastic small round cell tumor (EWS/WT1), Ewing's sarcoma (EWS/FLI1) and Wilms' tumor(WT1)[J]. PLoS One, 2013, 8(7): e68985. DOI:10.1371/journal.pone.0068985 |
| [41] |
YEUNG C, GIBSON A E, ISSAQ S H, et al. Targeting glycolysis through inhibition of lactate dehydrogenase impairs tumor growth in preclinical models of Ewing sarcoma[J]. Cancer Res, 2019, 79(19): 5060-5073. DOI:10.1158/0008-5472.CAN-19-0217 |
| [42] |
SARDESAI S D, THOMAS A, GALLAGHER C, et al. Inhibiting fatty acid synthase with omeprazole to improve efficacy of neoadjuvant chemotherapy in patients with operable TNBC[J]. Clin Cancer Res, 2021, 27(21): 5810-5817. DOI:10.1158/1078-0432.CCR-21-0493 |
| [43] |
KELLY W, DIAZ DUQUE A E, MICHALEK J, et al. Phase Ⅱ investigation of TVB-2640 (denifanstat) with bevacizumab in patients with first relapse high-grade astrocytoma[J]. Clin Cancer Res, 2023, 29(13): 2419-2425. DOI:10.1158/1078-0432.ccr-22-2807 |
| [44] |
MULLEN N J, SINGH P K. Nucleotide metabolism: a pan-cancer metabolic dependency[J]. Nat Rev Cancer, 2023, 23(5): 275-294. DOI:10.1038/s41568-023-00557-7 |
| [45] |
WU H L, GONG Y, JI P, et al. Targeting nucleotide metabolism: a promising approach to enhance cancer immunotherapy[J]. J Hematol Oncol, 2022, 15(1): 45. DOI:10.1186/s13045-022-01263-x |
| [46] |
SILIGAN C, BAN J, BACHMAIER R, et al. EWS-FLI1 target genes recovered from Ewing's sarcoma chromatin[J]. Oncogene, 2005, 24(15): 2512-2524. DOI:10.1038/sj.onc.1208455 |
| [47] |
ROBERT F, PELLETIER J. Perturbations of RNA helicases in cancer[J]. Wiley Interdiscip Rev RNA, 2013, 4(4): 333-349. DOI:10.1002/wrna.1163 |
| [48] |
ADAMSON P C, ZIMM S, RAGAB A H, et al. A phase Ⅱ trial of continuous-infusion 6-mercaptopurine for childhood solid tumors[J]. Cancer Chemother Pharmacol, 1990, 26(5): 343-344. DOI:10.1007/BF02897290 |
 2024, Vol. 45
2024, Vol. 45


