肾结石是泌尿系统常见且复发率极高的疾病,近年来我国的发病率超过10%[1],其中80%的肾结石主要成分是草酸钙[2]。随着微创外科技术的发展,肾结石的外科治疗手段日益丰富,但其治疗后10年的复发率高达50%[3]。当前预防肾结石发生和防止治疗后复发的手段仍非常缺乏,因此研究草酸钙结石的形成和发展机制十分必要。miRNA是一种短小的非编码RNA,通过与靶mRNA分子3' -非翻译区内的互补序列进行碱基配对,起到沉默mRNA表达和转录后调控的作用,据估计超过30%的蛋白质编码基因接受miRNA的调控[4]。然而泌尿系结石领域有关miRNA的研究非常少见。本研究通过对基因表达汇编(Gene Expression Omnibus,GEO)数据库中草酸钙肾结石大鼠肾组织的1个miRNA数据集(GSE72135)和1个mRNA数据集(GSE89028)进行联合分析,筛选出调控草酸钙肾结石形成的关键基因和miRNA,综合理解miRNA-mRNA调控网络机制,以期为草酸钙肾结石的诊治提供新思路。
1 资料和方法 1.1 数据集来源从GEO数据库中筛选出GSE72135(平台为GPL19117)和GSE89028(平台为GPL14797)2个数据集,前者为miRNA芯片数据集,包括3个对照和3个乙二醇喂养14 d诱导的草酸钙肾结石大鼠肾组织样本;后者为mRNA芯片数据集,共有6组24个样本(分组方法为对照、乙二醇诱导和乙二醇诱导同时加螺内酯,然后按诱导14 d和28 d分成2个亚组,每个亚组样本量为4个)。本研究选取其中乙二醇喂养14 d诱导的草酸钙肾结石大鼠肾组织及其对照的8个样本进行研究。
1.2 基因的差异表达分析利用GEO数据库自带的基于R语言的GEO2R在线分析工具,以P<0.05及|log2FC|>1[FC为差异表达倍数(fold change)]为阈值进行在线分析,筛选得到差异表达基因(differentially expressed gene,DEG)和差异表达miRNA(differentially expressed miRNA,DEM),并用R语言“pheatmap”包以热图的形式进行展示。
1.3 miRNA靶基因预测和调控基因筛选利用在线靶基因预测工具miRWalk(http://mirwalk.umm.uni-heidelberg.de/)分别预测数据集中上调和下调表达DEM的靶基因,然后根据miRNA负向调控mRNA的原理将预测得到的靶基因分别与下调和上调表达的DEG利用R语言“venn.diagram”包取交集,获得参与调控乙二醇诱导大鼠草酸钙肾结石形成的基因。
1.4 基因富集分析利用R语言“clusterProfiler”包对筛选出的草酸钙肾结石的调控基因进行基因本体(Gene Ontology,GO)功能富集分析和京都基因与基因组百科全书(Kyoto Encyclopedia of Genes and Genomes,KEGG)通路富集分析,以P<0.05作为筛选条件。
1.5 构建miRNA-mRNA调控网络把miRWalk预测得到的DEM和调控基因之间的关系对导入Cytoscape 3.9.1软件,构建miRNA-mRNA调控网络。
1.6 蛋白质-蛋白质相互作用(protein-protein interaction,PPI)网络分析和关键基因筛选将筛选出的调控基因导入String数据库,以关联度>0.4为阈值进行分析,结果导入Cytoscape 3.9.1软件对PPI网络进行可视化分析。应用cytohubba插件根据其提供的前5种算法即最大集团中心度(maximal clique centrality,MCC)、最大邻域分量(maximum neighborhood component,MNC)、最大邻域分量的密度(density of maximum neighborhood component,DMNC)、节点连接度(degree)和边缘渗透组件(edge percolated component,EPC),筛选出得分排名前20的基因,然后利用R语言“venn.diagram”包取交集,所得基因作为关键基因。
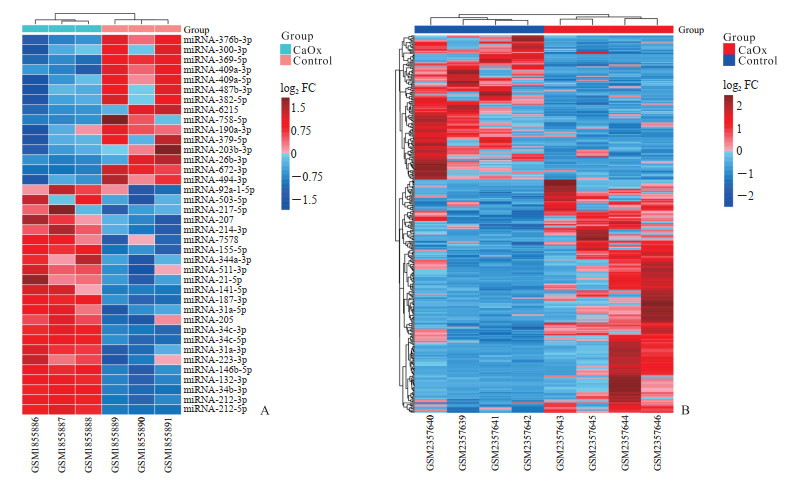
2 结果 2.1 DEM和DEG分析结果从GSE72135数据集中共筛选出DEM 38个,其中在草酸钙结石大鼠肾组织中上调表达的miRNA为23个,下调表达的miRNA为15个(图 1A)。从GSE89028数据集中共筛选出DEG 364个,其中上调表达基因225个,下调表达基因139个(图 1B)。
|
图 1 差异表达miRNA(A)和mRNA(B)的热图 Fig 1 Heatmap diagrams of differentially expressed miRNAs (A) and mRNAs (B) Red rectangles represent high expression, and blue ones represent low expression. Color depth represents the degree of difference. CaOx: Calcium oxalate; miRNA: MicroRNA; FC: Fold change. |
2.2 miRNA靶基因预测和调控基因筛选
利用在线靶基因预测工具miRWalk预测DEM的靶基因,15个低表达的miRNA预测到7 662个靶基因,然后根据miRNA负向调控mRNA原理与DEG中225个高表达的基因取交集,得到65个在草酸钙肾结石大鼠肾组织中上调表达的调控基因。23个高表达的miRNA预测到10 798个靶基因,与低表达的139个DEG取交集得到51个下调表达的调控基因。
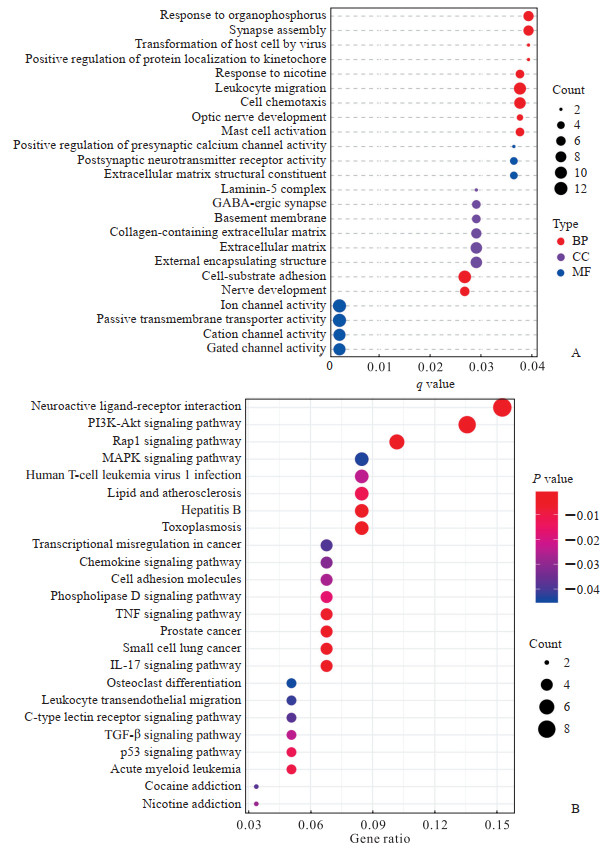
2.3 GO和KEGG富集分析应用“custerProfiler”包对筛选出的116个参与调控大鼠草酸钙肾结石形成的DEG进行GO功能富集分析,共富集到11条生物过程、6条细胞组分和7条分子功能条目,主要涉及着丝粒蛋白定位及其与微管的连接、阳离子通道及其激活、细胞外基质、神经突触的发育和功能等(图 2A)。KEGG通路富集分析共富集到24条信号通路,主要涉及PI3K-Akt、MAPK、TGF-β、神经活性配体-受体相互作用信号通路等(图 2B)。
|
图 2 116个调控基因的GO(A)和KEGG(B)富集分析 Fig 2 GO (A) and KEGG (B) enrichment analyses of 116 regulatory genes GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genome; BP: Biological process; CC: Cellular component; MF: Molecular function; GABA: γ-aminobutyric acid; PI3K: Phosphatidylinositol 3-kinase; Akt: Serine/threonine kinase; Rap1: Ras-related protein 1; MAPK: Mitogen-activated protein kinase; TNF: Tumor necrosis factor; IL-17: Interleukin 17; TGF-β: Transforming growth factor β. |
2.4 miRNA-mRNA调控网络
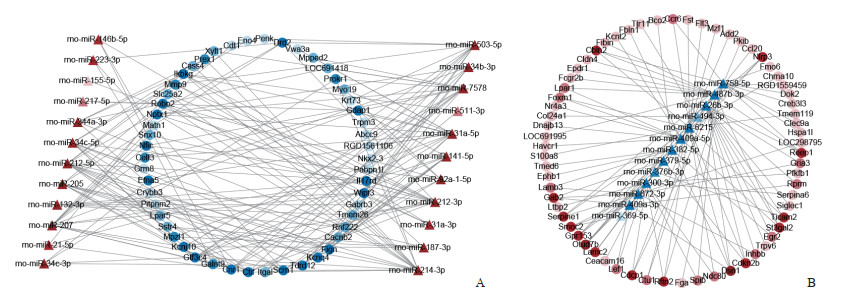
把利用miRWalk在线工具预测出的38个DEM和116个调控基因之间的调控关系对导入Cytoscapes 3.9.1软件进行可视化分析,获得包含152个节点和272条边的调控网络,其中下调表达的miRNA-203b-3p和miRNA-190a-3p未参与此调控网络。根据miRNA负向调控mRNA的原理,调控网络图分为上调表达miRNA调控低表达的mRNA(图 3A)和下调表达miRNA调控高表达的mRNA(图 3B)2部分。DEM中连接边最多的为miRNA-6215,调控了19个mRNA;其次为miRNA-758-5p和miRNA-214-3p,各调控了15个mRNA。而mRNA中连接边最多的为通用转录因子ⅢC亚基4(general transcription factor ⅢC subunit 4,Gtf3c4)和Ctif,分别接受10个和9个miRNA的调控。这些miRNA和mRNA可能在乙二醇诱导的大鼠草酸钙肾结石形成过程中起到关键作用。
|
图 3 miRNA-mRNA调控网络 Fig 3 miRNA-mRNA regulatory network A: The regulatory network of up-expressed miRNAs to low-expression mRNAs; B: The regulatory network of down-expressed miRNAs to high-expression mRNAs. The dots represent mRNAs, and triangles represent miRNAs. Red indicates up-regulated expression, blue represents down-regulated expression, and color depth reflects the number of connected edges of that node. miRNA, miR: MicroRNA; rno: Rattus norvegicus. |
2.5 PPI网络构建和关键基因筛选
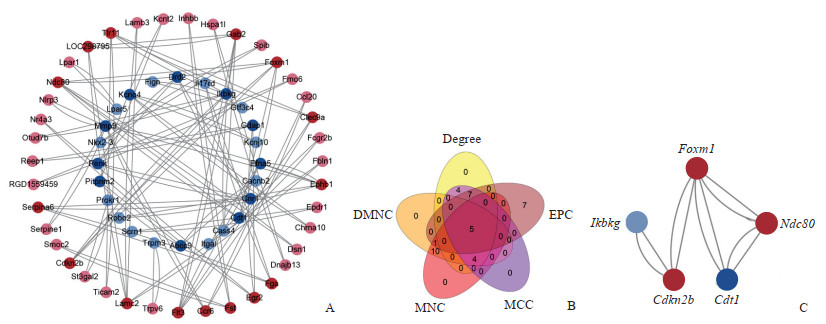
利用String数据库对116个调控基因进行PPI分析并将结果导入Cytoscape 3.9.1软件实现可视化,得到1个包含63个节点和100条边的PPI网络图(图 4A),应用cytohubba插件提供的前5种算法分别筛选出得分排名前20的基因(图 4B),然后利用“venn.diagram”包取交集得到5个关键基因:叉头框转录因子M1(forkhead box M1,Foxm1)、核分裂周期蛋白80(nuclear division cycle 80,Ndc80)、细胞周期蛋白依赖性激酶抑制因子2B(cyclin dependent kinase inhibitor 2B,Cdkn2b)、Cdc10依赖性转录因子1(Cdc10-dependent transcript 1,Cdt1)和B细胞编码κ轻链多肽抑制基因(inhibitor of nuclear factor κB kinase regulatory subunit γ,Ikbkg)(图 4C)。
|
图 4 调控基因的PPI网络图及关键基因模块 Fig 4 PPI network diagram of regulatory genes and hub genes A: PPI network diagram of regulatory genes. Circles represent genes and lines represent associations between genes. Deeper colors represent more connected nodes. B: Venn diagram of 5 groups of top 20 genes screened by different algorithms. C: The interaction network of hub genes. Red nodes indicate up-regulated expression, blue ones represents down-regulated expression, and color depth reflects the number of connected edges of that node. PPI: Protein-protein interaction; EPC: Edge percolated component; MCC: Maximal clique centrality; MNC: Maximum neighborhood component; DMNC: Density of maximum neighborhood component; Foxm1: Forkhead box M1; Ndc80: Nuclear division cycle 80; Cdkn2b: Cyclin dependent kinase inhibitor 2B; Cdt1: Cdc10-dependent transcript 1; Ikbkg: Inhibitor of nuclear factor κB kinase regulatory subunit γ. |
3 讨论
肾结石是泌尿系统的常见病和多发病,其发病率逐年上升,已成为慢性肾脏病和终末期肾病的重要原因之一。miRNA是一种内源性的短小的非编码RNA,通过构建沉默复合体负向调控下游基因的表达,据估计超过30%的蛋白质编码基因受miRNA的调控[4]。在本研究构建的miRNA-mRNA调控网络中,miRNA连接边最多的为miRNA-6215,调控了19个mRNA;其次为miRNA-758-5p和miRNA-214-3p,各调控了15个mRNA。miRNA-6215与miRNA-758-5p在数据集中为低表达,目前尚未检索到miRNA-6215相关研究,但miRNA-6215潜在靶向5个关键基因中的4个基因Foxm1、Ndc80、Cdkn2b和Ikbkg,具有进一步研究价值。已有研究表明miRNA-758-5p具有抑制胶质母细胞瘤生长和转移[5]、抑制脂肪吸收[6]、促进牙周膜干细胞的成骨和成牙分化[7]等功能。miRNA-214-3p在数据集中为高表达,其在肿瘤和非肿瘤领域都具有非常重要的生物功能。Zhou等[8]研究发现在顺铂诱导急性肾损伤过程中,miRNA-214-3p通过靶向谷胱甘肽过氧化物酶4加重肾小管上皮细胞的铁死亡,还有报道指出miRNA-214-3p参与巨噬细胞极化过程的调控[9],且铁死亡和巨噬细胞的极化均为影响草酸钙结石形成的重要因素[10-11]。在本研究中对调控基因进行KEGG富集分析得到的信号通路中,已有研究表明miRNA-214-3p参与调控PI3K/Akt、MAPK和TGF-β信号通路[12-14]。另外,肾小管上皮细胞损伤被认为是结石形成的起始点[15],而炎症反应和纤维化是结石形成和进展的重要机制[2],当前已有研究表明miRNA-223-3p和miRNA-21-5p参与草酸钙结晶性肾损伤中的调控,具有促进肾脏炎症和纤维化作用并可作为潜在的肾脏纤维化标志物[16]。此外,在本研究筛选出的DEM中,miRNA-214-3p、miRNA-503-5p、miRNA-21-5p、miRNA-223-3p、miRNA-146b-5p和miRNA-132-3p在另一项草酸钙肾结石大鼠芯片研究中也具有相同的表达趋势[17],为进一步研究miRNA在草酸钙肾结石形成过程中的功能提供了更为坚实的支撑。
本研究中,我们对调控基因进行了PPI网络分析,并且应用cytohubba插件得到了5个关键基因Foxm1、Ndc80、Cdkn2b、Cdt1和Ikbkg;其中Foxm1、Ndc80和Cdkn2b在草酸钙结石肾组织中高表达,而Cdt1和Ikbkg为低表达。Foxm1编码的蛋白属于叉头框(forkhead box,FOX)转录因子的一员。Foxm1在多种肿瘤中均为过表达,参与了Hanahan[18]所描述的所有癌症特征,干扰Foxm1的表达可以抑制肿瘤细胞增殖、转移、侵袭以及血管生成等生物过程,表明其可能成为抗癌治疗的靶目标[19-20]。研究发现Foxm1通过促进肾小管上皮细胞再生在急性肾损伤中发挥肾脏保护作用[21-22];同时另一项研究表明Foxm1促进单侧输尿管梗阻(unilateral ureteral obstruction,UUO)大鼠的肾脏纤维化[23];而Xie等[24]研究发现Foxm1通过调控Wnt蛋白家族多个成员的表达,激活Wnt/β-catenin通路促进UUO小鼠的肾脏纤维化,提示Foxm1可能在肾损伤的急性期发挥对肾脏的保护作用;而在损伤未能及时解除进入慢性损伤阶段时,Foxm1的持续高表达可导致过度修复并促进肾脏纤维化。在本研究筛选出的5个关键基因中,Foxm1差异表达程度最显著,同时接受处于调控网络中心位置的miRNA-6215的调控,因此miRNA-6215/Foxm1可作为草酸钙结石形成和发展机制研究的新方向。
Ndc80(人类的同源基因为Hec1)是构成Ndc80复合体的4个亚单位之一。Ndc80复合体在进化中高度保守,是纺锤体微管连接染色体着丝点的主要功能区域;而Ndc80的氨基端起着保持微观和着丝点的稳定性的作用,从而保证有丝分裂和子代染色体分离的顺利进行[25]。Ndc80基因在不同的恶性肿瘤中均过表达,常与不良临床预后相关,且诱导过度活化Ndc80基因会刺激癌细胞的增殖[25]。
Cdt1基因编码的蛋白质是细胞周期的调节中枢,并与DNA损伤修复有关。在染色体正常的情况下,Cdt1为DNA复制起点加载解旋酶复合物所必需,Cdt1耗竭使DNA复制许可活动失活,人工过表达Cdt1足以诱导基因组再复制[26]。同时Cdt1受到严格的调控,它仅在G1期存在于细胞核内,进入S期后迅速经E3泛素化途径水解,从而保证了在每个细胞周期只进行1次DNA复制[26-27]。在染色体受到损伤时,Cdt1被与损伤染色体上的核增殖抗原特异性结合的E3泛素连接酶CRL4Cdt2迅速招募到DNA损伤的位置并被蛋白酶水解,从而启动多种DNA修复途径[27]。Cdt1的表达在恶性肿瘤中通常升高,而此基因无效等位突变与Meier-Gorlin综合征相关[26, 28]。
Cdkn2b与抑癌基因Cdkn1a相邻,编码的蛋白为p15,目前认为Cdkn2b和Cdkn1a的表达和活性被骨髓细胞瘤病毒癌基因同源物(myelocytomatosis viral oncogene homolog,MYC)抑制从而参与MYC促肿瘤途径[29],然而Cdkn2b是否为抑癌基因尚需进一步研究。
Ikbkg基因又被称为NF-κB关键调节因子(Nemo)基因,其编码的蛋白质与IKKB-α和IKKB-β形成IκB激酶复合体,通过经典途径激活NF-κB通路调控炎症、免疫、细胞生存等通路的基因表达。人类Nemo基因突变以后引发一种X染色体连锁的称为Nemo综合征的联合免疫缺陷病[30]。Ikbkg在miRNA-mRNA调控网络中是连接边最多的关键基因,接受6个miRNA的靶向调控,其中miRNA-212-3p的连接边的数目和表达水平的差异程度都位于前列,且NF-κB信号通路在草酸钙结石形成过程中起着非常重要的促进作用[31-32],因此miRNA-212-3p/Ikbkg/NF-κB轴在结石形成过程中对炎症和免疫方面的调控作用具有进一步的研究价值。
5个关键基因中,Foxm1、Ndc80和Cdt1都能调节细胞周期并参与DNA损伤的修复[25-27, 33],当前研究显示DNA损伤可促进草酸钙结石形成[34],这也为进一步研究草酸钙结石形成的机制提供了新方向。
本研究通过对GEO数据库中筛选出的乙二醇诱导的大鼠草酸钙肾结石模型肾组织的miRNA和mRNA数据集进行综合数据分析,成功构建了miRNA-mRNA调控网络,验证了部分已阐明的草酸钙结石信号通路,同时也发现了尚未充分研究的信号通路,并筛选出了差异表达且处于关键调控位置的miRNA和关键基因,为草酸钙结石的防治提供了新思路和理论支撑。
| [1] |
WANG W, FAN J, HUANG G, et al. Prevalence of kidney stones in mainland China: a systematic review[J]. Sci Rep, 2017, 7: 41630. DOI:10.1038/srep41630 |
| [2] |
KHAN S R, CANALES B K, DOMINGUEZ-GUTIERREZ P R. Randall's plaque and calcium oxalate stone formation: role for immunity and inflammation[J]. Nat Rev Nephrol, 2021, 17(6): 417-433. DOI:10.1038/s41581-020-00392-1 |
| [3] |
WIGNER P, GRĘBOWSKI R, BIJAK M, et al. The molecular aspect of nephrolithiasis development[J]. Cells, 2021, 10(8): 1926. DOI:10.3390/cells10081926 |
| [4] |
WANG B, WU B, LIU J, et al. Analysis of altered microRNA expression profiles in proximal renal tubular cells in response to calcium oxalate monohydrate crystal adhesion: implications for kidney stone disease[J]. PLoS One, 2014, 9(7): e101306. DOI:10.1371/journal.pone.0101306 |
| [5] |
LIU J, JIANG J, HUI X, et al. miR-758-5p suppresses glioblastoma proliferation, migration and invasion by targeting ZBTB20[J]. Cell Physiol Biochem, 2018, 48(5): 2074-2083. DOI:10.1159/000492545 |
| [6] |
LI B R, XIA L Q, LIU J, et al. miR-758-5p regulates cholesterol uptake via targeting the CD363'UTR[J]. Biochem Biophys Res Commun, 2017, 494(1/2): 384-389. DOI:10.1016/j.bbrc.2017.09.150 |
| [7] |
YAN C, LI N, XIAO T, et al. Extracellular vesicles from the inflammatory microenvironment regulate the osteogenic and odontogenic differentiation of periodontal ligament stem cells by miR-758-5p/LMBR1/BMP2/4 axis[J]. J Transl Med, 2022, 20: 208-215. DOI:10.1186/s12967-022-03412-9 |
| [8] |
ZHOU J, XIAO C, ZHENG S, et al. MicroRNA-214-3p aggravates ferroptosis by targeting GPX4 in cisplatin-induced acute kidney injury[J]. Cell Stress Chaperones, 2022, 27(4): 325-336. DOI:10.1007/s12192-022-01271-3 |
| [9] |
PENG L Y, LI B B, DENG K B, et al. MicroRNA-214-3p facilitates M2 macrophage polarization by targeting GSK3B[J]. Kaohsiung J Med Sci, 2022, 38(4): 347-356. DOI:10.1002/kjm2.12487 |
| [10] |
TAGUCHI K, OKADA A, UNNO R, et al. Macrophage function in calcium oxalate kidney stone formation: a systematic review of literature[J]. Front Immunol, 2021, 12: 673690. DOI:10.3389/fimmu.2021.673690 |
| [11] |
SONG Q, LIAO W, CHEN X, et al. Oxalate activates autophagy to induce ferroptosis of renal tubular epithelial cells and participates in the formation of kidney stones[J]. Oxid Med Cell Longev, 2021, 2021: 6630343. DOI:10.1155/2021/6630343 |
| [12] |
LV X, YANG H, ZHONG H, et al. Osthole exhibits an antitumor effect in retinoblastoma through inhibiting the PI3K/AKT/mTOR pathway via regulating the hsa_circ_0007534/miR-214-3p axis[J]. Pharm Biol, 2022, 60(1): 417-426. DOI:10.1080/13880209.2022.2032206 |
| [13] |
WAN H, TIAN Y, ZHAO J, et al. LINC00665 targets miR-214-3p/MAPK1 axis to accelerate hepatocellular carcinoma growth and Warburg effect[J]. J Oncol, 2021, 2021: 9046798. DOI:10.1155/2021/9046798 |
| [14] |
YANG F, QIN Y, LV J, et al. Silencing long non-coding RNA Kcnq1ot1 alleviates pyroptosis and fibrosis in diabetic cardiomyopathy[J]. Cell Death Dis, 2018, 9: 1000. DOI:10.1038/s41419-018-1029-4 |
| [15] |
WANG Z, ZHANG Y, ZHANG J, et al. Recent advances on the mechanisms of kidney stone formation (Review)[J]. Int J Mol Med, 2021, 48(2): 149. DOI:10.3892/ijmm.2021.4982 |
| [16] |
GHOLAMINEJAD A, ABDUL TEHRANI H, GHOLAMI FESHARAKI M. Identification of candidate microRNA biomarkers in renal fibrosis: a meta-analysis of profiling studies[J]. Biomarkers, 2018, 23(8): 713-724. DOI:10.1080/1354750X.2018.1488275 |
| [17] |
LAN C, CHEN D, LIANG X, et al. Integrative analysis of miRNA and mRNA expression profiles in calcium oxalate nephrolithiasis rat model[J]. BioMed Res Int, 2017, 2017: 8306736. DOI:10.1155/2017/8306736 |
| [18] |
HANAHAN D. Hallmarks of cancer: new dimensions[J]. Cancer Discov, 2022, 12(1): 31-46. DOI:10.1158/2159-8290.CD-21-1059 |
| [19] |
时佳琪, 刘超, 张艳桥. FoxM1调节网络——抗癌新靶点[J]. 现代肿瘤医学, 2019, 27(6): 1059-1063. DOI:10.3969/j.issn.1672-4992.2019.06.038 |
| [20] |
LIU C, BARGER C J, KARPF A R. FOXM1:a multifunctional oncoprotein and emerging therapeutic target in ovarian cancer[J]. Cancers, 2021, 13(12): 3065. DOI:10.3390/cancers13123065 |
| [21] |
CHANG-PANESSO M, KADYROV F F, LALLI M, et al. FOXM1 drives proximal tubule proliferation during repair from acute ischemic kidney injury[J]. J Clin Investig, 2019, 129(12): 5501-5517. DOI:10.1172/jci125519 |
| [22] |
SINHA S, DWIVEDI N, WOODGETT J, et al. Glycogen synthase kinase-3β inhibits tubular regeneration in acute kidney injury by a FoxM1-dependent mechanism[J]. FASEB J, 2020, 34(10): 13597-13608. DOI:10.1096/fj.202000526rr |
| [23] |
WANG Y, ZHOU Q, TANG R, et al. FoxM1 inhibition ameliorates renal interstitial fibrosis by decreasing extracellular matrix and epithelial-mesenchymal transition[J]. J Pharmacol Sci, 2020, 143(4): 281-289. DOI:10.1016/j.jphs.2020.05.007 |
| [24] |
XIE H, MIAO N, XU D, et al. FoxM1 promotes Wnt/β-catenin pathway activation and renal fibrosis via transcriptionally regulating multi-Wnts expressions[J]. J Cellular Molecular Medi, 2021, 25(4): 1958-1971. DOI:10.1111/jcmm.15948 |
| [25] |
USTINOV N B, KORSHUNOVA A V, GUDIMCHUK N B. Protein complex NDC80:properties, functions, and possible role in pathophysiology of cell division[J]. Biochemistry (Mosc), 2020, 85(4): 448-462. |
| [26] |
ZHANG H. Regulation of DNA replication licensing and re-replication by Cdt1[J]. Int J Mol Sci, 2021, 22(10): 5195. DOI:10.3390/ijms22105195 |
| [27] |
KANELLOU A, GIAKOUMAKIS N N, PANAGOPOULOS A, et al. The licensing factor Cdt1 links cell cycle progression to the DNA damage response[J]. Anticancer Res, 2020, 40(5): 2449-2456. DOI:10.21873/anticanres.14214 |
| [28] |
BURRAGE L C, CHARNG W L, ELDOMERY M K, et al. De novo GMNN mutations cause autosomal-dominant primordial dwarfism associated with Meier-Gorlin syndrome[J]. Am J Hum Genet, 2015, 97(6): 904-913. DOI:10.1016/j.ajhg.2015.11.006 |
| [29] |
GARCÍ A-GUTIÉ RREZ L, DELGADO M D, LEÓ N J. MYC oncogene contributions to release of cell cycle brakes[J]. Genes, 2019, 10(3): 244. DOI:10.3390/genes10030244 |
| [30] |
刘洋, 胡坚. NEMO综合征研究进展[J]. 中国实用儿科杂志, 2019, 34(11): 949-953. DOI:10.19538/j.ek2019110617 |
| [31] |
MING S, TIAN J, MA K, et al. Oxalate-induced apoptosis through ERS-ROS-NF-κB signalling pathway in renal tubular epithelial cell[J]. Mol Med, 2022, 28(1): 88. DOI:10.1186/s10020-022-00494-5 |
| [32] |
AN L, WU W, LI S, et al. Escherichia coli aggravates calcium oxalate stone formation via PPK1/flagellin-mediated renal oxidative injury and inflammation[J]. Oxid Med Cell Longev, 2021, 2021: 9949697. DOI:10.1155/2021/9949697 |
| [33] |
PAL S, KOZONO D, YANG X, et al. Dual HDAC and PI3K inhibition abrogates NFkappaB- and FOXM1-mediated DNA damage response to radiosensitize pediatric high-grade gliomas[J]. Cancer Res, 2018, 78(14): 4007-4021. DOI:10.1158/0008-5472.CAN-17-3691 |
| [34] |
KITTIKOWIT W, WAIWIJIT U, BOONLA C, et al. Increased oxidative DNA damage seen in renal biopsies adjacent stones in patients with nephrolithiasis[J]. Urolithiasis, 2014, 42(5): 387-394. DOI:10.1007/s00240-014-0676-x |
 2023, Vol. 44
2023, Vol. 44


