强直性脊柱炎(ankylosing spondylitis,AS)是一种病因未明、具有致残性、以骶髂关节和脊柱附着点炎症为主要症状的疾病,多见于男性,其发病年龄趋于年轻化[1]。AS是脊柱外科和风湿免疫科的研究重点之一,现有研究显示AS主要与遗传、环境、免疫和内分泌等病理因素密切相关[2-4]。目前临床中仍缺乏用于早期诊断和治疗AS的生物标志物,因此探索AS相关的重要靶点十分必要。近年来,随着生物信息学技术的发展,许多疾病相关的全基因组测序成为研究热点。有研究采用环形RNA芯片技术检测AS患者外周血单核细胞中环形RNA的表达谱,发现环形RNA或许可成为AS诊断和进展观察的重要标志物[5]。单细胞测序技术在AS疾病中的应用进一步揭示了AS进展的潜在机制[6]。本研究拟通过检索基因表达汇编(Gene Expression Omnibus,GEO)公共数据库中AS相关基因芯片数据,分析、筛选差异表达基因,并对差异表达基因进行功能、信号通路富集分析及蛋白质-蛋白质相互作用(protein-protein interaction,PPI)网络分析,挖掘AS诊断的新型标志物,以期为AS的早期诊治提供新思路。
1 资料和方法 1.1 数据收集从美国国立生物技术信息中心(National Center of Biotechnology Information,NCBI)的GEO数据库中检索AS相关的芯片数据,并下载获得芯片GSE25101数据集[7],平台文件均为GPL6947(Illumina HumanHT-12 V3.0 expression beadchip)。GSE25101数据集中包含健康志愿者和AS患者外周血全血样本各16例。
1.2 差异表达基因筛选通过R语言4.0 Bioconductor项目中的limma数据分析包比较健康志愿者和AS患者外周血中的基因表达改变来识别差异表达基因。差异表达基因的筛选标准均设定为校正后P<0.05和差异倍数≥1.25或≤0.8,根据结果绘制火山图,观察上调和下调基因的表达改变,然后通过聚类分析进一步明确基因间和样本间的分布关系。
1.3 基因功能与信号通路富集分析利用注视、可视化和集成发现数据库(Database for Annovation,Visualization,and Integrated Discovery;DAVID)(https://david.ncifcrf.gov/)对差异表达基因进行基因本体(Gene Ontology,GO)功能和京都基因与基因组百科全书(Kyoto Encyclopedia of Genes and Genomes,KEGG)信号通路分析,仅错误发现率(false discovery rate,FDR)<0.05具有统计学意义。
1.4 PPI网络构建与关键基因筛选将得到的差异表达基因导入在线数据库String(http://string-db.org/)构建PPI网络,然后利用Cytoscape 3.7.1软件中的MCODE和cytoHubba插件筛选PPI网络中最为显著的蛋白质模块获取关键基因。
1.5 关键基因对AS的诊断价值分析利用R语言4.0绘制ROC曲线并计算AUC值,评估关键基因对AS的诊断价值。检验水准(α)为0.05。
2 结果 2.1 差异表达基因分析结果从健康志愿者和AS患者外周血全血样本中共筛选出差异表达基因187个,其中上调基因96个、下调基因91个。聚类分析结果显示两组样本质量合格,差异表达基因的热图见图 1。
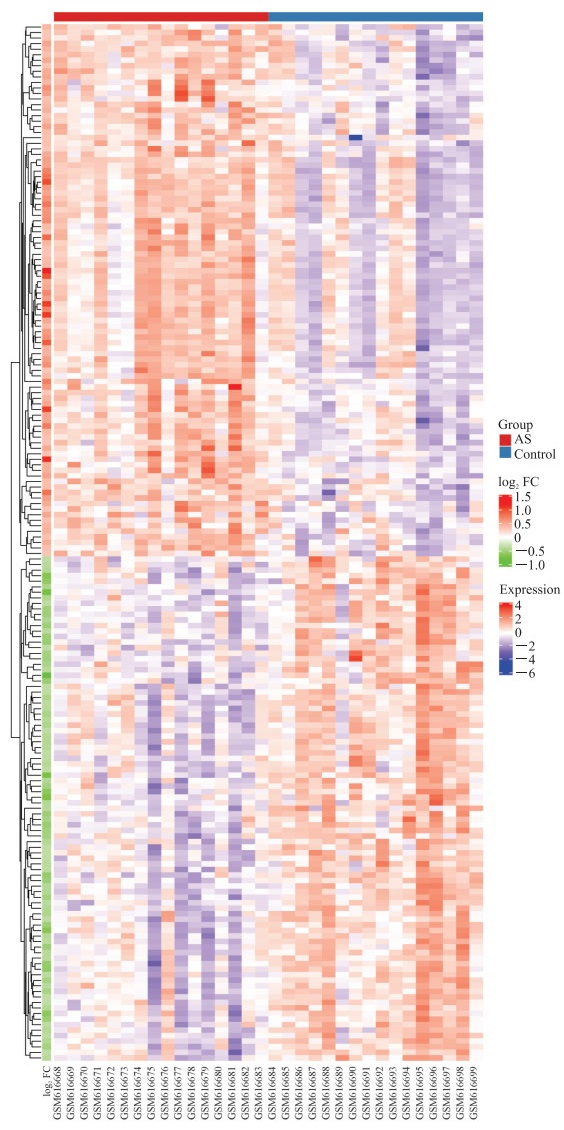
|
图 1 AS患者与健康志愿者差异表达基因的热图 Fig 1 Heatmap of differentially expressed genes between AS patients and healthy controls AS: Ankylosing spondylitis; FC: Fold change. |
2.2 差异表达基因的GO功能和KEGG信号通路分析
GO功能分析结果显示,差异表达基因参与的生物学过程主要为核糖核苷一磷酸代谢过程、核苷一磷酸代谢过程和嘌呤核糖核苷一磷酸代谢过程等(图 2A),细胞组分主要为线粒体呼吸链、呼吸链和线粒体膜部分等,而分子功能则以氢离子跨膜转运活性和晚期糖基化终末产物受体(receptor of advanced glycation end product,RAGE)结合等为主。KEGG信号通路分析结果显示,差异表达基因主要富集于非酒精性脂肪肝、亨廷顿病和氧化磷酸化等通路(图 2B)。

|
图 2 AS患者与健康志愿者差异表达基因的GO生物过程和KEGG信号通路分析 Fig 2 GO biological process and KEGG signaling pathway analyses of differentially expressed genes between AS patients and healthy controls A: The biological process of GO function analysis; B: The enrichment result of the KEGG signaling pathway. GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; ATP: Adenosine triphosphate. |
2.3 PPI网络构建和蛋白质模块分析
将187个差异表达基因导入在线数据库String,按照组合得分>0.400的标准并隐藏未参与构建PPI网络的蛋白,将输出结果导入Cytoscape 3.7.1软件,结果显示蛋白质网络共有134个节点(蛋白)和311条边(蛋白之间的相互联系)(图 3A)。利用MCODE插件进行蛋白质模块分析,获得最为显著模块和10个关键基因(图 3B):模块由10个节点和42条边组成,这10个节点分别为NADH: 泛醌氧化还原酶亚基S4(NADH: ubiquinone oxidoreductase subunit S4,NDUFS4)、细胞色素c氧化酶亚基5B(cytochrome c oxidase subunit 5B,COX5B)、泛醇-细胞色素c还原酶结合蛋白(ubiquinol-cytochrome c reductase binding protein,UQCRB)、细胞色素c氧化酶亚基7B(cytochrome c oxidase subunit 7B,COX7B)、泛醇-细胞色素c还原酶铰链蛋白(ubiquinol-cytochrome c reductase hinge protein,UQCRH)、ATP合成酶F1亚基ε假基因2(ATP synthase F1 subunit epsilon pseudogene 2,ATP5EP2)、ATP合成酶H+转运线粒体F0复合体亚基F6(ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F6;ATP5J)、细胞色素c氧化酶亚基6A1(cytochrome c oxidase subunit 6A1,COX6A1)、NADH:泛醌氧化还原酶亚基B3(NADH: ubiquinone oxidoreductase subunitB3,NDUFB3)和细胞色素c氧化酶亚基7A2(cytochrome c oxidase subunit 7A2,COX7A2)。再使用cytoHubba插件对10个关键基因进行评分排序,获得最为显著的5个关键基因,即ATP5J、COX7A2、NDUFB3、UQCRB和UQCRH(图 3C)。

|
图 3 PPI网络分析和关键基因获取 Fig 3 PPI network analysis and hub gene acquisition A: PPI network analysis. Cytoscape 3.7.1 software was used to visualize the interaction between differentially expressed genes; B: The most significant module in PPI network was obtained through MCODE plug-in analysis, which contained 10 hub genes; C: Hub genes were obtained by cytoHubba plug-in (the redder the node color is, the higher the score is). PPI: Protein-protein interaction. |
2.4 关键基因对AS的诊断价值分析
利用ROC曲线评估上述5个关键基因诊断AS的灵敏度和特异度。如图 4A所示,ATP5J(AUC=0.859)、NDUFB3(AUC=0.852)、UQCRB(AUC=0.840)、COX7A2(AUC=0.820)和UQCRH(AUC=0.805)的AUC值均>0.600,AUC值越接近1,表明这5个基因诊断疾病的灵敏度和特异度越高。联合5个关键基因构建疾病诊断模型(图 4B),发现联合诊断的AUC=0.875,进一步明确这5个关键基因可作为AS的诊断标志物。
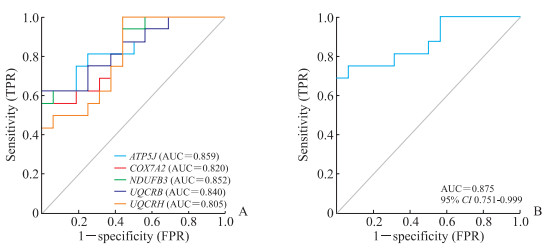
|
图 4 ROC曲线评估关键基因对AS的诊断能力 Fig 4 ROC curves for evaluating the diagnostic performance of hub genes for AS A: The diagnostic performance of the 5 hub genes; B: The diagnostic performance of the model constructed by the 5 hub genes. ROC: Receiver operating characteristic; AS: Ankylosing spondylitis; TPR: True positive rate; FPR: False positive rate; ATP5J: Adenosine triphosphate synthase, H+transporting, mitochondrial F0 complex, subunit F6; COX7A2: Cytochrome c oxidase subunit 7A2; NDUFB3: NADH: ubiquinone oxidoreductase subunit B3; UQCRB: Ubiquinol-cytochrome c reductase binding protein; UQCRH: Ubiquinol-cytochrome c reductase hinge protein; AUC: Area under curve; CI: Confidence interval. |
3 讨论
AS是临床上的常见病,影响患者的生活质量,严重者甚至可导致残疾,因此通过生物信息学技术探索AS的发病机制,挖掘疾病相关的生物标志物,对早期诊治AS具有重要意义。
本研究通过生物信息学技术分析了健康人和AS患者外周血样本的基因表达情况,筛选出差异表达基因187个,其中包含上调基因96个和下调基因91个。GO功能富集分析结果显示,差异表达基因主要参与氧化磷酸化代谢等生物学过程。AS患者存在明显的氧化应激损伤,氧化应激损伤与疾病活动度呈正相关[8]。有研究表明抗炎治疗AS可明显改善各类脂质和酶类的活性,这或许将成为AS治疗的方向之一[9]。应用黄芩清热除痹胶囊治疗AS的临床试验结果表明,黄芩清热除痹胶囊可以改善AS患者的临床症状,其机制可能与上调叉头框蛋白O3a(forkhead box protein O3a,FOXO3a)表达,减轻AS患者的免疫炎症和氧化应激有关[10]。KEGG信号通路富集分析结果显示,差异表达基因主要富集于氧化磷酸化等信号通路。PPI网络分析和ROC曲线分析进一步明确5个关键基因ATP5J、NDUFB3、UQCRB、COX7A2和UQCRH可作为AS的诊断标志物。
ATP5J是一种连接ATP合酶F0和F1组分的蛋白,参与肾癌、肝癌和消化道肿瘤等多种疾病进展[11-13],提示氧化磷酸化途径或许为AS的潜在治疗方向。COX7A2是细胞色素c氧化酶(cytochrome c oxidase,COX)的组成部分之一,而COX是线粒体呼吸链的末端酶并参与合成生命活动所需物质[14],且COX相关代谢途径包括ATP合成、呼吸电子传递和产热等,与本研究的GO功能富集分析结果一致。研究发现COX7A2表达升高与阿尔茨海默病患者海马Aβ斑块负荷显著相关,可能是阿尔茨海默病重要的风险因素之一[15]。NDUFB3基因编码线粒体膜呼吸链还原型烟酰胺腺嘌呤二核苷酸(reduced nicotinamide adenine dinucleotide,NADH)脱氢酶的一个附属亚基,可诱导NOD样受体热蛋白结构域相关蛋白3(NOD-like receptor thermal protein domain associated protein 3,NLRP3)激活和细胞焦亡[16]。UQCRB基因编码泛素-细胞色素c氧化还原酶复合物的一个亚基,减弱UQCRB的表达可抑制线粒体活性氧生成,进而阻断缺氧诱导因子激活和血管内皮生长因子受体2信号转导[17-18]。UQCRH分布于细胞核和线粒体,主要参与线粒体氧化磷酸化,其异常高表达可能导致细胞活性氧产生,从而促进癌基因的表达和肿瘤的发生、发展[19],该基因目前已被识别作为多种疾病的治疗靶点和预后指标[20-21]。
筛选疾病相关的诊断标志物可为早期诊治疾病提供方向。Kyritsis等[22]通过分析鉴定脊髓损伤急性期患者外周血基因差异表达情况,获取了与脊髓损伤严重程度和预后相关的标志物。Yu等[23]采用色谱质谱法筛选AS组和非AS组的髋关节韧带样本中差异表达蛋白,发现髓过氧化物酶或许可成为AS诱发髋关节病变相关的重要标志物。采用全血样本筛选疾病相关诊断标志物较获取组织样本的方法更便捷。研究发现lncRNA ITSN1-2[24]和镁离子依赖的蛋白磷酸酶1A(protein phosphatase magnesium-dependent 1A,PPM1A)[25]的表达水平与经过TNF-α抑制剂治疗的AS患者疗效相关,具有一定的临床价值。García-Salinas等[26]通过队列研究评估人体白细胞抗原B27(human leukocyte antigen B27,HLA-B27)作为轴性脊柱炎诊断标志物的效能,结果显示HLA-B27诊断轴性脊柱炎疾病特异度良好,但灵敏度较低。本研究通过生物信息学分析方法筛选出AS患者外周血诊断标志物ATP5J、NDUFB3、UQCRB、COX7A2和UQCRH,结果显示这5个基因联合应用可明显提高AS的诊断效力。这些标志物亦可作为AS治疗靶点,或许可成为未来AS非手术治疗的一种重要方式,减少组织工程修复和外科手术等治疗方式的风险。
综上所述,本研究通过生物信息学技术分析了GEO数据芯片,识别出ATP5J、NDUFB3、UQCRB、COX7A2和UQCRH可作为AS诊断的标志物,为进一步明确AS的潜在病理机制及早期临床诊治提供了理论参考,并为AS靶向治疗药物的研发提供了方向。
| [1] |
CROSSFIELD S S R, MARZO-ORTEGA H, KINGSBURY S R, PUJADES-RODRIGUEZ M, CONAGHAN P G. Changes in ankylosing spondylitis incidence, prevalence and time to diagnosis over two decades[J/OL]. RMD Open, 2021, 7: e001888. DOI: 10.1136/rmdopen-2021-001888.
|
| [2] |
SCHLOSSTEIN L, TERASAKI P I, BLUESTONE R, PEARSON C M. High association of an HL-A antigen, W27, with ankylosing spondylitis[J]. N Engl J Med, 1973, 288: 704-706. DOI:10.1056/NEJM197304052881403 |
| [3] |
WU Y F, REN M L, YANG R, LIANG X J, MA Y C, TANG Y, et al. Reduced immunomodulation potential of bone marrow-derived mesenchymal stem cells induced CCR4+CCR6+ Th/Treg cell subset imbalance in ankylosing spondylitis[J/OL]. Arthritis Res Ther, 2011, 13: R29. DOI: 10.1186/ar3257.
|
| [4] |
KEBAPCILAR L, BILGIR O, ALACACIOGLU A, YILDIZ Y, TAYLAN A, GUNAYDIN R, et al. Impaired hypothalamo-pituitary-adrenal axis in patients with ankylosing spondylitis[J]. J Endocrinol Invest, 2010, 33: 42-47. DOI:10.1007/BF03346548 |
| [5] |
TANG Y P, ZHANG Q B, DAI F, LIAO X, DONG Z R, YI T, et al. Circular RNAs in peripheral blood mononuclear cells from ankylosing spondylitis[J]. Chin Med J (Engl), 2021, 134: 2573-2582. DOI:10.1097/CM9.0000000000001815 |
| [6] |
XU H X, YU H Y, LIU L X, WU H W, ZHANG C T, CAI W X, et al. Integrative single-cell RNA-seq and ATAC-seq analysis of peripheral mononuclear cells in patients with ankylosing spondylitis[J/OL]. Front Immunol, 2021, 12: 760381. DOI: 10.3389/fimmu.2021.760381.
|
| [7] |
PIMENTEL-SANTOS F M, LIGEIRO D, MATOS M, MOURÃO A F, COSTA J, SANTOS H, et al. Whole blood transcriptional profiling in ankylosing spondylitis identifies novel candidate genes that might contribute to the inflammatory and tissue-destructive disease aspects[J/OL]. Arthritis Res Ther, 2011, 13: R57. DOI: 10.1186/ar3309.
|
| [8] |
SOLMAZ D, KOZACI D, SARIİ, TAYLAN A, ÖNEN F, AKKOÇ N, et al. Oxidative stress and related factors in patients with ankylosing spondylitis[J]. Eur J Rheumatol, 2016, 3: 20-24. DOI:10.5152/eurjrheum.2015.0031 |
| [9] |
CZÓKOLYOVÁ M, PUSZTAI A, VÉGH E, HORVÁTH Á, SZENTPÉTERI A, HAMAR A, et al. Changes of metabolic biomarker levels upon one-year anti-TNF-α therapy in rheumatoid arthritis and ankylosing spondylitis: associations with vascular pathophysiology[J/OL]. Biomolecules, 2021, 11: 1535. DOI: 10.3390/biom11101535.
|
| [10] |
黄旦, 刘健, 纵瑞凯, 万磊. 黄芩清热除痹胶囊激活PPARγ介导AMPK/FOXO3a信号通路改善强直性脊柱炎患者氧化应激[J]. 中国中药杂志, 2020, 45: 451-456. |
| [11] |
BAI B J, XIE B B, PAN Z Y, SHAN L N, ZHAO J P, ZHU H B. Identification of candidate genes and long non-coding RNAs associated with the effect of ATP5J in colorectal cancer[J]. Int J Oncol, 2018, 52: 1129-1138. |
| [12] |
YANG W W, LU Y, XU Y C, XU L Z, ZHENG W, WU Y Y, et al. Estrogen represses hepatocellular carcinoma (HCC) growth via inhibiting alternative activation of tumor-associated macrophages (TAMs)[J]. J Biol Chem, 2012, 287: 40140-40149. |
| [13] |
BJERREGAARD H, PEDERSEN S, KRISTENSEN S R, MARCUSSEN N. Reference genes for gene expression analysis by real-time reverse transcription polymerase chain reaction of renal cell carcinoma[J]. Diagn Mol Pathol, 2011, 20: 212-217. DOI:10.1097/PDM.0b013e318212e0a9 |
| [14] |
TIMÓN-GÓMEZ A, NYVLTOVÁ E, ABRIATA L A, VILA A J, HOSLER J, BARRIENTOS A. Mitochondrial cytochrome c oxidase biogenesis: recent developments[J]. Semin Cell Dev Biol, 2018, 76: 163-178. DOI:10.1016/j.semcdb.2017.08.055 |
| [15] |
BI R, ZHANG W, ZHANG D F, XU M, FAN Y, HU Q X, et al. Genetic association of the cytochrome c oxidase-related genes with Alzheimer's disease in Han Chinese[J]. Neuropsychopharmacology, 2018, 43: 2264-2276. |
| [16] |
CHUNG I C, CHEN L C, TSANG N M, CHUANG W Y, LIAO T C, YUAN S N, et al. Mitochondrial oxidative phosphorylation complex regulates NLRP3 inflammasome activation and predicts patient survival in nasopharyngeal carcinoma[J]. Mol Cell Proteomics, 2020, 19: 142-154. DOI:10.1074/mcp.RA119.001808 |
| [17] |
JUNG H J, CHO M, KIM Y, HAN G, KWON H J. Development of a novel class of mitochondrial ubiquinol-cytochrome c reductase binding protein (UQCRB) modulators as promising antiangiogenic leads[J]. J Med Chem, 2014, 57: 7990-7998. DOI:10.1021/jm500863j |
| [18] |
JUNG H J, KIM K H, KIM N D, HAN G, KWON H J. Identification of a novel small molecule targeting UQCRB of mitochondrial complex Ⅲ and its anti-angiogenic activity[J]. Bioorg Med Chem Lett, 2011, 21: 1052-1056. |
| [19] |
GAO F, LIU Q C, LI G P, DONG F, QIU M L, LV X T, et al. Identification of ubiquinol cytochrome c reductase hinge (UQCRH) as a potential diagnostic biomarker for lung adenocarcinoma[J/OL]. Open Biol, 2016, 6: 150256. DOI: 10.1098/rsob.150256.
|
| [20] |
PARK E R, KIM S B, LEE J S, KIM Y H, LEE D H, CHO E H, et al. The mitochondrial hinge protein, UQCRH, is a novel prognostic factor for hepatocellular carcinoma[J]. Cancer Med, 2017, 6: 749-760. |
| [21] |
YU H T, LIU Y C, HE B R, HE T, CHEN C Y, HE J H, et al. Platelet biomarkers for a descending cognitive function: a proteomic approach[J/OL]. Aging Cell, 2021, 20: e13358. DOI: 10.1111/acel.13358.
|
| [22] |
KYRITSIS N, TORRES-ESPÍN A, SCHUPP P G, HUIE J R, CHOU A, DUONG-FERNANDEZ X, et al. Diagnostic blood RNA profiles for human acute spinal cord injury[J/OL]. J Exp Med, 2021, 218: e20201795. DOI: 10.1084/jem.20201795.
|
| [23] |
YU C J, ZHAN X L, LIANG T, CHEN L Y, ZHANG Z D, JIANG J, et al. Mechanism of hip arthropathy in ankylosing spondylitis: abnormal myeloperoxidase and phagosome[J/OL]. Front Immunol, 2021, 12: 572592. DOI: 10.3389/fimmu.2021.572592.
|
| [24] |
LI M W, ZHOU X J. Long noncoding RNA intersectin 1-2 gradually declines during adalimumab treatment, and its reduction correlates with treatment efficacy in patients with ankylosing spondylitis[J]. Inflammopharmacology, 2021, 29: 1371-1378. |
| [25] |
LEE J S, LEE E J, LEE J H, HONG S C, LEE C K, YOO B, et al. Autoantibodies against protein phosphatase magnesium-dependent 1A as a biomarker for predicting radiographic progression in ankylosing spondylitis treated with anti-tumor necrosis factor agents[J/OL]. J Clin Med, 2020, 9: 3968. DOI: 10.3390/jcm9123968.
|
| [26] |
GARCÍA-SALINAS R, RUTA S, CHICHANDE J T, MAGRI S. The role of HLA-B27 in argentinian axial spondyloarthritis patients[J/OL]. J Clin Rheumatol, 2022, 28: e619-e622. DOI: 10.1097/RHU.0000000000001763.
|
 2022, Vol. 43
2022, Vol. 43


