2. 海军军医大学(第二军医大学)海军特色医学中心妇产科,上海 200433
2. Department of Obstetrics and Gynecology, Naval Medical Center, Naval Medical University (Second Military Medical University), Shanghai 200433, China
卵巢癌的发病率和病死率在女性恶性肿瘤中均排名第8位[1]。早期缺乏有效的诊断方法与晚期高复发率是造成卵巢癌高病死率的主要原因[2]。因此,寻找有效的肿瘤标志物并研究其在卵巢癌发生、发展中的作用对卵巢癌的诊断、预防和治疗具有重要意义。本研究通过生物信息学技术寻找与卵巢癌预后相关的关键基因,为卵巢癌的治疗提供新的靶点。
1 资料和方法 1.1 基因芯片数据获取从基因表达汇编(Gene Expression Omnibus,GEO)数据库下载GSE18520和GSE14407数据集中的相关数据,前者包括53个肿瘤样本和10个正常卵巢样本,后者包括12个肿瘤样本和12个正常卵巢样本,所用平台均为GPL570(Affymetrix Human Genome U133 Plus 2.0 Array)。从基因型-组织表达(Genotype-Tissue Expression,GTEx)数据库中获得88例卵巢正常组织的数据,从癌症基因组图谱(The Cancer Genome Atlas,TCGA)数据库中获得379例卵巢癌组织的数据。
1.2 差异表达基因获取使用R 3.6.2软件limma包筛选卵巢癌与正常组织之间的差异表达基因,以P < 0.05和|log2FC|>2[FC为差异倍数(fold change)]为筛选标准。使用维恩图获得GEO数据库GSE18520、GSE14407数据集及TCGA、GTEx数据库中重叠的基因。
1.3 基因功能和通路富集分析利用R 3.6.2软件clusterProfiler包进行基因本体(gene ontology,GO)和京都基因与基因组百科全书(Kyoto Encyclopedia of Genes and Genomes,KEGG)富集分析,设定P < 0.05为差异有统计学意义。
1.4 核心基因筛选使用STRING数据库(http://string-db.org/)进行蛋白质-蛋白质相互作用(protein-protein interaction,PPI)网络构建。通过Cytoscape软件[3]进一步分析PPI网络,利用cytoHubba插件[4]根据Matthews相关系数(Matthews correlation coefficient,MCC)算法筛选核心基因。
1.5 核心基因验证利用基因表达谱交互分析(Gene Expression Profiling Interactive Analysis,GEPIA)数据库(http://gepia.cancer-pku.cn)[5]验证获得的核心基因在卵巢癌中的表达情况。通过人类蛋白质图谱(Human Protein Atlas,HPA)数据库(https://www.proteinatlas.org)[6]分析核心基因在卵巢癌和正常组织中的蛋白表达。在Kaplan-Meier Plotter数据库(https://www.kmplot.com)中对核心基因进行生存分析。
2 结果 2.1 差异表达基因分析通过GEO数据库GSE18520和GSE14407数据集获得了211个共同的正常卵巢样本与卵巢肿瘤组织的差异表达基因,其中53个表达上调,158个表达下调。通过GTEx结合TCGA数据库进行分析,获得2 253个差异表达基因,其中1 017个表达上调,1 236个表达下调。进一步分析发现,TCGA数据库中有69个差异表达基因与GEO数据库GSE18520和GSE14407数据集的重叠基因匹配,其中35个表达上调、34个表达下调。对69个基因进行GO富集分析发现,生物过程主要富集于间充质细胞分化和泌尿生殖系统中,细胞组分主要富集于纺锤体,分子功能主要富集于受体-配体活性等;KEGG富集分析显示,信号通路主要富集在ABC转运体、视黄醇代谢和Wnt信号通路(图 1)。
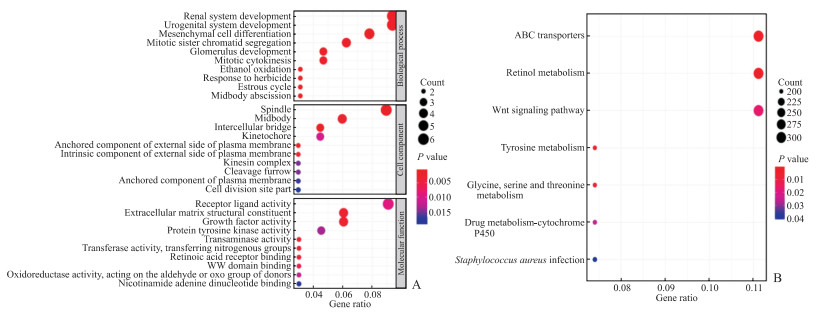
|
图 1 卵巢癌差异表达基因的GO和KEGG富集分析 Fig 1 GO and KEGG enrichment analyses of differentially expressed genes of ovarian cancer A: GO functional enrichment analysis; B: KEGG pathway enrichment analysis. GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes. |
2.2 核心基因获取和验证
在STRING中构建了69个差异基因的PPI网络,包括26个节点和70个边,经计算获得10个核心基因,其中BUB1有丝分裂检查点丝氨酸/苏氨酸激酶B(BUB1 mitotic checkpoint serine/threonine kinase B,BUB1B)基因在卵巢癌中研究颇多,故本文中不做具体分析;其余9个基因分别为细胞周期依赖性激酶亚基蛋白2(cyclin dependent-kinase subunit protein 2,CKS2)、母体胚胎亮氨酸拉链激酶(maternal embryonic leucine zipper kinase,MELK)、序列相似性83家族蛋白成员D(family with sequence similarity 83,member D;FAM83D)、中心体相关蛋白55(centrosomal protein 55,CEP55)、叉头框蛋白M1(forkhead box M1,FOXM1)、中心体相关激酶2(NIMA related kinase 2,NEK2)、驱动蛋白家族成员20A(kinesin family member 20A,KIF20A)、TTK蛋白激酶(TTK protein kinase,TTK)、驱动蛋白家族成员4A(kinesin family member 4A,KIF4A)。GEPIA数据库分析表明,这9个核心基因在卵巢癌组织中高表达(图 2),并且NEK2表达水平随卵巢癌分期的增高而降低(图 3)。通过HPA数据库获取卵巢癌患者的临床免疫组织化学染色样本,结果显示卵巢癌组织中CEP55和KIF20A蛋白呈高表达(图 4)。
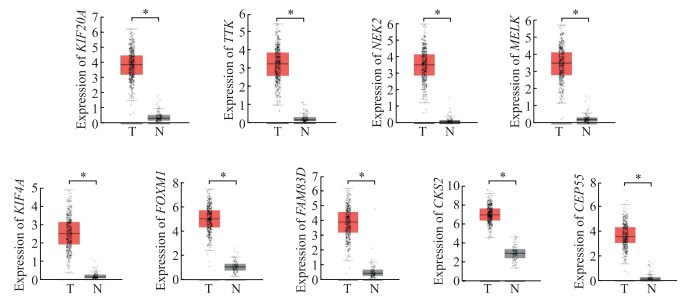
|
图 2 GEPIA数据库中卵巢正常组织与卵巢癌组织核心基因的表达 Fig 2 Expression of hub genes in normal and ovarian cancer tissues in GEPIA database *P < 0.05. n=426 in ovarian cancer tissue (T) group, n=88 in normal ovarian tissue (N) group. GEPIA: Gene Expression Profile Interaction Analysis; KIF20A: Kinesin family member 20A; TTK: TTK protein kinase; NEK2: NIMA related kinase 2; MELK: Maternal embryonic leucine zipper kinase; KIF4A: Kinesin family member 4A; FOXM1: Forkhead box M1; FAM83D: Family with sequence similarity 83, member D; CKS2: Cyclin dependent-kinase subunit protein 2; CEP55: Centrosomal protein 55. |
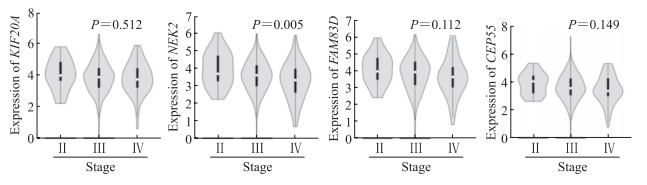
|
图 3 GEPIA数据库中核心基因与卵巢癌分期的关系 Fig 3 Relationship between hub genes and ovarian cancer stages in GEPIA database GEPIA: Gene Expression Profile Interaction Analysis; KIF20A: Kinesin family member 20A; NEK2: NIMA related kinase 2; FAM83D: Family with sequence similarity 83, member D; CEP55: Centrosomal protein 55. |
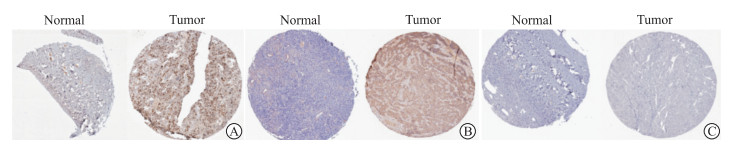
|
图 4 HPA数据库中正常卵巢组织与卵巢癌组织核心基因的蛋白表达情况 Fig 4 Protein expression of hub genes in normal ovarian tissues and ovarian cancer tissues in HPA database A: KIF20A; B: CEP55; C: NEK2. Immunohistochemistry (40×). HPA: Human Protein Atlas; KIF20A: Kinesin family member 20A; CEP55: Centrosomal protein 55; NEK2: NIMA related kinase 2. |
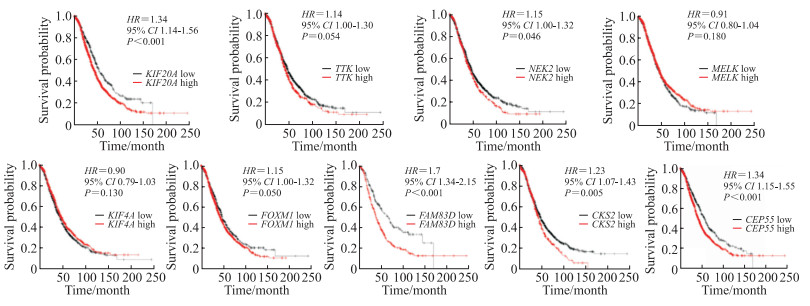
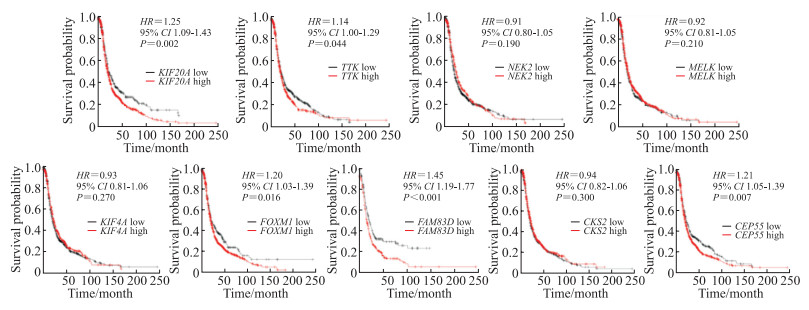
Kaplan-Meier Plotter数据库生存分析结果显示,CEP55、CKS2、FAM83D、KIF20A和NEK2高表达的卵巢癌患者总生存期比低表达的患者短(图 5),而CEP55、FOXM1、FAM83D、KIF20A和TTK高表达的卵巢癌患者无进展生存期比低表达的患者短(图 6)。
|
图 5 Kaplan-Meier Plotter数据库中核心基因表达与卵巢癌患者总生存期的关系 Fig 5 Association of expression of hub genes with overall survival of ovarian cancer patients in Kaplan-Meier Plotter database KIF20A: Kinesin family member 20A; TTK: TTK protein kinase; NEK2: NIMA related kinase 2; MELK: Maternal embryonic leucine zipper kinase; KIF4A: Kinesin family member 4A; FOXM1: Forkhead box M1; FAM83D: Family with sequence similarity 83, member D; CKS2: Cyclin dependent-kinase subunit protein 2; CEP55: Centrosomal protein 55; HR: Hazard ratio; CI: Confidence interval |
|
图 6 Kaplan-Meier Plotter数据库中核心基因表达与卵巢癌患者无进展生存期的关系 Fig 6 Association of expression of hub genes with progress free survival of ovarian cancer patients in Kaplan-Meier Plotter database KIF20A: Kinesin family member 20A; TTK: TTK protein kinase; NEK2: NIMA related kinase 2; MELK: Maternal embryonic leucine zipper kinase; KIF4A: Kinesin family member 4A; FOXM1: Forkhead box M1; FAM83D: Family with sequence similarity 83, member D; CKS2: Cyclin dependent-kinase subunit protein 2; CEP55: Centrosomal protein 55; HR: Hazard ratio; CI: Confidence interval. |
3 讨论
虽然卵巢癌的治疗方法和手术方式已经有所改进,但晚期卵巢癌患者由于诊断困难,治疗结果和预后仍然很差,探索与卵巢癌预后相关的基因非常必要。本研究从卵巢癌组织和正常卵巢组织芯片数据中得到69个差异表达基因(其中上调基因35个,下调基因34个)。GO富集分析显示,在生物学过程中,差异基因主要集中在间充质细胞分化及泌尿生殖系统中。大量研究表明上皮-间质转化(epithelial-mesenchymal transition,EMT)在胚胎发育中发挥了关键作用,同时也参与了肿瘤的进展和转移[7-9];上皮钙黏蛋白(epithelial cadherin,E-cadherin)与神经钙黏蛋白(neural cadherin,N-cadherin)是EMT中的重要分子,研究发现,E-cadherin表达增多与N-cadherin表达下降可降低卵巢癌细胞的侵袭能力[9]。KEGG富集分析发现,差异表达基因主要富集在ABC转运体、视黄醇代谢和Wnt信号通路。卵巢癌是一种常见的易出现化学治疗耐药的实体肿瘤,既往研究发现ABC转运蛋白可致癌症的多药耐药,包括多柔比星、依托泊苷和长春新碱等[10],而ABC转运蛋白在卵巢癌中的研究并不多见,这也为研究卵巢癌化学治疗耐药提供了新的思路。视黄醇代谢已被证明与乳腺癌和胆囊癌有关[11]。在胚胎和成人组织稳态中,Wnt/β-catenin通路调节细胞增殖、极性、存活和干细胞命运,Wnt信号通路异常与肿瘤的发生等多种病理过程有关[12-13],越来越多的研究证明Wnt信号通路影响卵巢癌的血管生成、转移、化学治疗耐药和免疫逃逸等诸多方面[14-15]。
本研究通过PPI网络分析筛选出9个核心基因。在卵巢癌组织中这9个核心基因的表达均高于卵巢正常组织。生存分析结果显示,CEP55、CKS2、FAM83D、KIF20A和NEK2高表达的患者总生存期较短,并且其中CEP55、FAM83D和KIF20A也与患者的无进展生存期有关。此外,NEK2与卵巢癌分期相关。近年研究发现CEP55参与调控PI3K/AKT通路和癌细胞干细胞化[16-18]。临床研究发现CEP55在乳腺癌、前列腺癌、肾癌、甲状腺癌等多种癌症中高表达[19-20],高表达的CEP55蛋白与非小细胞肺癌的不良预后相关[21]。本研究发现,CEP55高表达的卵巢癌患者的总生存期和无进展生存期比低表达的患者短。CKS2属于细胞周期依赖蛋白激酶亚基家族,参与细胞周期调控[22]。研究表明,CKS2表达下调可抑制结直肠癌患者肿瘤细胞增殖、促进凋亡[23]。本研究发现CKS2的高表达预示着卵巢癌患者总生存期较差,但不影响患者的无进展生存期。FAM83D可能通过抑制抑癌因子含F框和WD重复域蛋白7(F-box and WD repeat domain containing 7,FBXW7)在乳腺癌中发挥致癌作用[24],同时可以促进肝癌增殖和侵袭[25]。本研究结果表明,FAM83D高表达的卵巢癌患者总生存期和无进展生存期较差。KIF20A与细胞增殖、迁移和化学治疗耐药有关。许多研究证实,KIF20A在肺癌[26]、胃癌[27]、肝癌[28]等恶性肿瘤中高表达,然而,其与卵巢癌的相关性尚不清楚。在本研究中,KIF20A高表达卵巢癌患者总生存期和无进展生存期较差。NEK2是宫颈癌组织中过表达的丝氨酸/苏氨酸激酶,与肿瘤分期和淋巴结转移有关[29]。本研究发现NEK2与卵巢癌的预后和卵巢癌的肿瘤分期有关,同时其高表达也与卵巢癌患者预后不良有关。以上这些基因与不同癌症的发生、发展密切相关,但在卵巢癌中的研究并不多见,因其与卵巢癌的预后密切相关,因此后期进行体内及体外实验的验证是非常必要的。
本研究结果显示,CEP55、CKS2、FAM83D、KIF20A和NEK2在卵巢癌组织中的mRNA水平高于正常卵巢组织;但根据HPA结果,只有CEP55和KIF20A的蛋白水平在肿瘤组织中高于正常组织;这可能与蛋白质的某些修饰有关,但具体机制尚不清楚。
综上所述,本研究通过对多数据库进行分析,发现CEP55、CKS2、FAM83D、KIF20A、NEK2、FOXM1、TTK与卵巢癌患者预后相关;此外,NEK2与卵巢癌分期相关。本研究仅为基于多数据库的分析结果,因此后期需要从细胞与动物实验方面进行以上基因的验证,其具体作用机制也有待进一步探索。
| [1] |
SUNG H, FERLAY J, SIEGEL R L, LAVERSANNE M, SOERJOMATARAM I, JEMAL A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71: 209-249. DOI:10.3322/caac.21660 |
| [2] |
XU Z Z, ZHOU Y, CAO Y X, DINH T L A, WAN J, ZHAO M. Identification of candidate biomarkers and analysis of prognostic values in ovarian cancer by integrated bioinformatics analysis[J/OL]. Med Oncol, 2016, 33: 130. DOI: 10.1007/s12032-016-0840-y.
|
| [3] |
SU G, MORRIS J H, DEMCHAK B, BADER G D. Biological network exploration with Cytoscape 3[J/OL]. Curr Protoc Bioinformatics, 2014, 47: 8.13.1-8.1324. DOI: 10.1002/0471250953.bi0813s47.
|
| [4] |
CHIN C H, CHEN S H, WU H H, HO C W, KO M T, LIN C Y. cytoHubba: identifying hub objects and subnetworks from complex interactome[J/OL]. BMC Syst Biol, 2014, 8: S11. DOI: 10.1186/1752-0509-8-S4-S11.
|
| [5] |
TANG Z F, LI C W, KANG B X, GAO G, LI C, ZHANG Z M. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses[ J/OL]. Nucleic Acids Res, 2017, 45: W98-W102. DOI: 10.1093/nar/gkx247.
|
| [6] |
UHLEN M, ZHANG C, LEE S, SJÖSTEDT E, FAGERBERG L, BIDKHORI G, et al. A pathology atlas of the human cancer transcriptome[J/OL]. Science, 2017, 357: eaan2507. DOI: 10.1126/science.aan2507.
|
| [7] |
DUAN H Y, YAN Z Q, CHEN W, WU Y, HAN J S, GUO H Y, et al. TET1 inhibits EMT of ovarian cancer cells through activating Wnt/β-catenin signaling inhibitors DKK1 and SFRP2[J]. Gynecol Oncol, 2017, 147: 408-417. DOI:10.1016/j.ygyno.2017.08.010 |
| [8] |
IWATSUKI M, MIMORI K, YOKOBORI T, ISHI H, BEPPU T, NAKAMORI S, et al. Epithelialmesenchymal transition in cancer development and its clinical significance[J]. Cancer Sci, 2010, 101: 293-299. DOI:10.1111/j.1349-7006.2009.01419.x |
| [9] |
ROSSO M, MAJEM B, DEVIS L, LAPYCKYJ L, BESSO M J, LLAURADÓ M, et al. E-cadherin: a determinant molecule associated with ovarian cancer progression, dissemination and aggressiveness[J/OL]. PLoS One, 2017, 12: e0184439. DOI: 10.1371/journal.pone.0184439.
|
| [10] |
AMAWI H, SIM H M, TIWARI A K, AMBUDKAR S V, SHUKLA S. ABC transporter-mediated multidrugresistant cancer[J]. Adv Exp Med Biol, 2019, 1141: 549-580. |
| [11] |
CHEN A C, GUO X, DERGUINI F, GUDAS L J. Human breast cancer cells and normal mammary epithelial cells: retinol metabolism and growth inhibition by the retinol metabolite 4-oxoretinol[J]. Cancer Res, 1997, 57: 4642-4651. |
| [12] |
CLEVERS H. Wnt/β-catenin signaling in development and disease[J]. Cell, 2006, 127: 469-480. DOI:10.1016/j.cell.2006.10.018 |
| [13] |
POLAKIS P. The many ways of Wnt in cancer[J]. Curr Opin Genet Dev, 2007, 17: 45-51. DOI:10.1016/j.gde.2006.12.007 |
| [14] |
AREND R C, LONDOÑO-JOSHI A I, STRAUGHN J M Jr, BUCHSBAUM D J. The Wnt/β-catenin pathway in ovarian cancer: a review[J]. Gynecol Oncol, 2013, 131: 772-779. DOI:10.1016/j.ygyno.2013.09.034 |
| [15] |
NAGARAJ A B, JOSEPH P, KOVALENKO O, SINGH S, ARMSTRONG A, REDLINE R, et al. Critical role of Wnt/β-catenin signaling in driving epithelial ovarian cancer platinum resistance[J]. Oncotarget, 2015, 6: 23720-23734. DOI:10.18632/oncotarget.4690 |
| [16] |
CHEN C H, LU P J, CHEN Y C, FU S L, WU K J, TSOU A P, et al. FLJ10540-elicited cell transformation is through the activation of PI3-kinase/AKT pathway[J]. Oncogene, 2007, 26: 4272-4283. DOI:10.1038/sj.onc.1210207 |
| [17] |
WANG G Z, LIU M N, WANG H J, YU S, JIANG Z F, SUN J H, et al. Centrosomal protein of 55 regulates glucose metabolism, proliferation and apoptosis of glioma cells via the Akt/mTOR signaling pathway[J]. J Cancer, 2016, 7: 1431-1440. DOI:10.7150/jca.15497 |
| [18] |
KUO T C, CHEN C T, BARON D, ONDER T T, LOEWER S, ALMEIDA S, et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity[J]. Nat Cell Biol, 2011, 13: 1214-1223. DOI:10.1038/ncb2332 |
| [19] |
SHIRAISHI T, TERADA N, ZENG Y, SUYAMA T, LUO J, TROCK B, et al. Cancer/testis antigens as potential predictors of biochemical recurrence of prostate cancer following radical prostatectomy[J/OL]. J Transl Med, 2011, 9: 153. DOI: 10.1186/1479-5876-9-153.
|
| [20] |
JONES J, OTU H, SPENTZOS D, KOLIA S, INAN M, BEECKEN W D, et al. Gene signatures of progression and metastasis in renal cell cancer[J]. Clin Cancer Res, 2005, 11: 5730-5739. DOI:10.1158/1078-0432.CCR-04-2225 |
| [21] |
JIANG C, ZHANG Y, LI Y, LU J, HUANG Q, XU R, et al. High CEP55 expression is associated with poor prognosis in non-small-cell lung cancer[J]. Onco Targets Ther, 2018, 11: 4979-4990. DOI:10.2147/OTT.S165750 |
| [22] |
CHEN R, FENG C, XU Y. Cyclin-dependent kinaseassociated protein CKS2 is associated with bladder cancer progression[J]. J Int Med Res, 2011, 39: 533-540. DOI:10.1177/147323001103900222 |
| [23] |
YU M H, LUO Y, QIN S L, WANG Z S, MU Y F, ZHONG M. Up-regulated CKS2 promotes tumor progression and predicts a poor prognosis in human colorectal cancer[J]. Am J Cancer Res, 2015, 5: 2708-2718. |
| [24] |
WANG Z R, LIU Y Y, ZHANG P J, ZHANG W G, WANG W J, CURR K, et al. FAM83D promotes cell proliferation and motility by downregulating tumor suppressor gene FBXW7[J]. Oncotarget, 2013, 4: 2476-2486. DOI:10.18632/oncotarget.1581 |
| [25] |
LIAO W J, LIU W L, LIU X, YUAN Q, OU Y, QI Y, et al. Upregulation of FAM83D affects the proliferation and invasion of hepatocellular carcinoma[J]. Oncotarget, 2015, 6: 24132-24147. DOI:10.18632/oncotarget.4432 |
| [26] |
ZHAO X, ZHOU L L, LI X Y, NI J, CHEN P, MA R, et al. Overexpression of KIF20A confers malignant phenotype of lung adenocarcinoma by promoting cell proliferation and inhibiting apoptosis[J]. Cancer Med, 2018, 7: 4678-4689. DOI:10.1002/cam4.1710 |
| [27] |
YAN G R, ZOU F Y, DANG B L, ZHANG Y, YU G C, LIU X, et al. Genistein-induced mitotic arrest of gastric cancer cells by downregulating KIF20A, a proteomics study[J]. Proteomics, 2012, 12: 2391-2399. DOI:10.1002/pmic.201100652 |
| [28] |
GASNEREAU I, BOISSAN M, MARGALL-DUCOS G, C O U C H Y G, W E N D U M D, B O U R G A I N - GUGLIELMETTI F, et al. KIF20A mRNA and its product MKlp2 are increased during hepatocyte proliferation and hepatocarcinogenesis[J]. Am J Pathol, 2012, 180: 131-140. DOI:10.1016/j.ajpath.2011.09.040 |
| [29] |
XU T, ZENG Y, SHI L, YANG Q, CHEN Y, WU G, et al. Targeting NEK2 impairs oncogenesis and radioresistance via inhibiting the Wnt1/β-catenin signaling pathway in cervical cancer[J/OL]. J Exp Clin Cancer Res, 2020, 39: 183. DOI: 10.1186/s13046-020- 01659-y.
|
 2022, Vol. 43
2022, Vol. 43


