结肠癌是严重威胁我国居民生命健康的高发消化道恶性肿瘤之一,目前建立的包括根治性手术、放疗、化疗、分子靶向治疗、免疫治疗等在内的综合治疗手段极大地改善了结肠癌患者预后,但是仍有很大一部分患者出现治疗后复发、转移,最终导致死亡率高[1-2]。导致这种治疗困境的根本原因主要在于结肠癌的肿瘤异质性较大[3-5],分子调控机制尤为精细,单一、片面的研究结论无法解释所有现象,从转录水平探索结肠癌的潜在转录调控机制十分必要。由于染色质结构的特殊性,大部分基因组DNA处于与核小体紧密结合缠绕的状态,无法被反式因子接近、结合进而调控;小部分DNA因无核小体包裹而裸露在外,这些区域被称为染色质开放区。染色质开放区在基因表达调控方面起着核心节点的作用,是各种调控因子对基因进行调控的必经之路[6-7],对其进行深入研究有助于了解结肠癌发生、发展过程中调控机制的变化。
染色质开放性高通量测序(assay for transposase-accessible chromatin using sequencing, ATAC-seq)技术是研究染色质开放状态的新技术,弥补了MNase-Seq、FAIRE-Seq和DNAse-Seq等研究方法的许多不足[8-9],在基因调控领域具有极高的应用价值。本研究利用癌症基因组图谱(The Cancer Genome Atlas, TCGA)[10]数据库的ATAC-seq数据,对结肠癌进展过程中的相关功能和信号通路改变进行分析,从染色质开放状态层面探讨结肠癌的通路及转录调控机制,为结肠癌的机制研究提供参考及线索。
1 资料和方法 1.1 结核癌组织样本ATAC-seq和转录本测序(RNA sequencing, RNA-seq)数据的下载及预处理从TCGA数据库(https://cancergenome.nih.gov/)下载结肠腺癌ATAC-seq数据,包括测序peaks在各样本分布的log2 (norm)标准化数据和TCGA_identifier_mapping样本名称数据,对两者进行整合,得到通用样本名称的ATAC-seq数据peaks的标准化数据。然后下载结肠腺癌RNA-seq数据,获得每千个碱基的转录每百万映射读取的片段数(fragments per kilobase of exon model per million mapped fragments, FPKM)。提取同时检测ATAC-seq和RNA-seq的组织样本测序数据,用于后续分析。
1.2 ATAC-seq数据的质量控制使用R 3.5.3软件,先加载karyoploteR包,对结肠癌ATAC-seq数据峰值(peaks)进行染色体覆盖度可视化;然后加载ChIPseeker包、TxDb.Hsapiens.UCSC.hg38.knownGene包、org.Hs.eg.db包、clusterProfiler包,对ATAC-seq数据peaks的位置特征进行归纳分类并可视化,进一步绘制维恩图展示结果。
1.3 结肠癌全部ATAC-seq数据peaks注释基因基因本体(Gene Ontology, GO)功能和京都基因与基因组百科全书(Kyoto Encyclopedia of Genes and Genomes, KEGG)通路富集分析使用R 3.5.3软件,加载ChIPseeker包、TxDb.Hsapiens.UCSC.hg38.knownGene包、org.Hs.eg.db包、clusterProfiler包、ggplot2包,对结肠癌全部ATAC-seq数据peaks进行基因注释,并对其进行GO功能富集分析及KEGG通路富集分析。
1.4 结肠癌关键基因的表达与其启动子区peaks的相关性分析利用TCGA数据库中同时检测ATAC-seq和RNA-seq的结肠癌组织样本测序数据,计算结肠癌关键基因肿瘤抑制基因APC(APC regulator of Wnt signaling pathway, APC)[11]、Kirsten大鼠肉瘤病毒癌基因(Kirsten rat sarcoma viral oncogene, KRAS)[12]、v-raf小鼠肉瘤病毒癌基因同源物B1(v-raf murine sarcoma viral oncogene homolog B1, BRAF)[13]的表达(RNA-seq的FPKM数据)与注释到该基因启动子区的ATAC-seq数据peaks的Pearson相关系数。
1.5 TNM分期Ⅲ+Ⅳ期与Ⅰ+Ⅱ期差异开放peaks注释基因的KEGG通路富集分析将结肠癌ATAC-seq数据的peaks按照TNM分期分成Ⅰ+Ⅱ期和Ⅲ+Ⅳ期两组。使用R 3.5.3软件,加载limma包,利用Wilcoxon秩和检验筛选两者之间的差异peaks,筛选条件为log2|FC|>1[FC为差异倍数(fold change)]且校正P<0.05,并加载ChIPseeker包、TxDb.Hsapiens.UCSC.hg38.knownGene包、org.Hs.eg.db包、clusterProfiler包对差异peaks进行启动子区域基因注释。最后按照TNM分期Ⅲ+Ⅳ期相对Ⅰ+Ⅱ期上调和下调分为2类,分别对注释基因进行KEGG通路富集分析。
2 结果 2.1 组织样本的一般情况共有36个结肠癌组织样本纳入本研究(经质量控制,无排除样本),均含有ATAC-seq数据和RNA-seq数据。其中有17个TNM分期Ⅰ+Ⅱ期和19个Ⅲ+Ⅳ期结肠癌组织样本。检测的ATAC-seq数据peaks为122 872个,RNA-seq数据FPKM为19 658个。
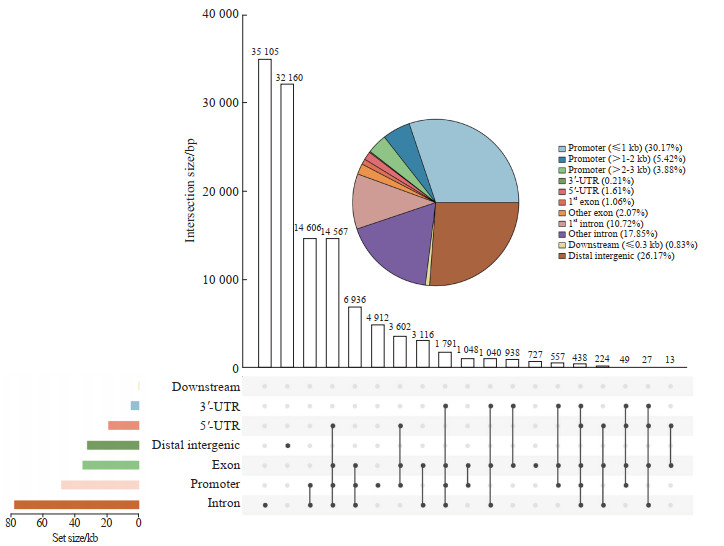
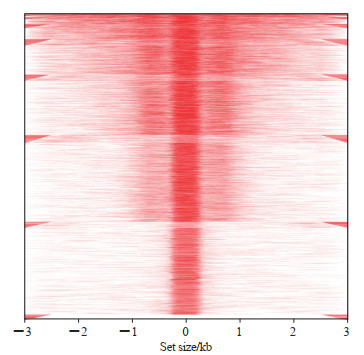
2.2 结肠癌ATAC-seq数据质量控制情况由图 1可见,ATAC-seq数据中检测到的peaks在各个染色体分布较为均匀。由图 2可见,位于启动子区域的peaks占比最高(距离转录起始位点≤1 kb、>1~2 kb、>2~3 kb者分别占30.17%、5.42%、3.88%),其次为远端基因间区(26.17%),符合染色质开放区的2种主要类型的分布,即基因上游的启动子和远端的调控元件(增强子和沉默子)。维恩图分析显示,染色质开放区peaks可以符合多种位置特征,间接解释了生物调控的多样性与复杂性。由图 3可见,染色质开放区peaks大多位于转录起始位点附近,符合染色质开放性的特征,证实了测序数据的可靠性与真实性。

|
图 1 结肠癌ATAC-seq数据peaks覆盖度图 Fig 1 Covered length of ATAC-seq peaks of colon cancer The abscissa represents the chromosome position, and the red band represents the standardized value of peaks. ATAC-seq: Assay for transposase-accessible chromatin using sequencing; Chr: Chromosome. |
|
图 2 结肠癌ATAC-seq数据peaks位置特征饼图及维恩图 Fig 2 Pie chart and Venn diagram of location characteristics of ATAC-seq peaks of colon cancer ATAC-seq: Assay for transposase-accessible chromatin using sequencing; UTR: Untranslated region. |
|
图 3 结肠癌ATAC-seq数据peaks距离转录起始位点分布热图 Fig 3 Heat map of distance from peaks to transcription initiation sites in ATAC-seq data of colon cancer The negative value represents the 5' end of transcription initiation site, the positive value represents the 3' end of transcription initiation site, and the red line represents peaks. ATAC-seq: Assay for transposase-accessible chromatin using sequencing. |
2.3 结肠癌全部ATAC-seq数据peaks注释基因KEGG通路富集分析结果
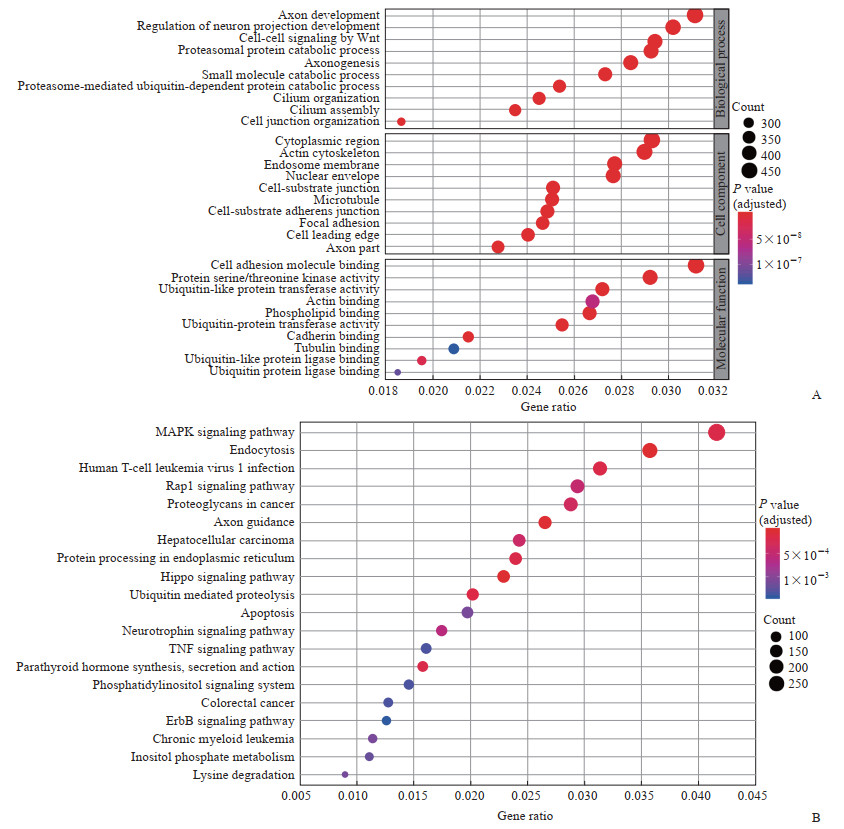
结肠癌样本中大量癌症相关GO功能和KEGG信号通路出现显著富集,包括Wnt信号通路的细胞间信号转导、细胞黏附分子结合相关功能、结直肠癌信号通路、表皮生长因子受体(epidermal growth factor receptor, ErbB)信号通路、MAPK信号通路、细胞凋亡、TNF信号通路等(图 4),体现了结肠癌发病过程中异常增殖信号通路大量活化、调控稳态逐渐失衡的过程。
|
图 4 结肠癌ATAC-seq数据peaks注释基因GO功能(A)和KEGG通路(B)富集分析 Fig 4 GO function (A) and KEGG pathway (B) enrichment analyses of annotated genes of colon cancer ATAC-seq peaks ATAC-seq: Assay for transposase-accessible chromatin using sequencing; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; MAPK: Mitogen-activated protein kinase; TNF: Tumor necrosis factor; ErbB: Epidermal growth factor receptor. |
2.4 3个结肠癌关键基因的表达与其启动子区ATAC-seq数据peaks的相关性
APC、KRAS、BRAF基因启动子区ATAC-seq数据peaks均与其RNA-seq的FPKM呈正相关(APC-chr5:112737575- 112738074:r=0.419,P=0.011;KRAS-chr12: 25250266-25250765:r=0.437,P=0.008;KRAS-chr12:25384622-25385121:r=0.42,P=0.011;BRAF-chr7:140924032-140924531:r=0.337,P=0.045;BRAF-chr7:140924750-140925249:r=0.359,P=0.032),提示染色质开放性可能通过直接影响结肠癌的关键基因参与结肠癌发生、发展。
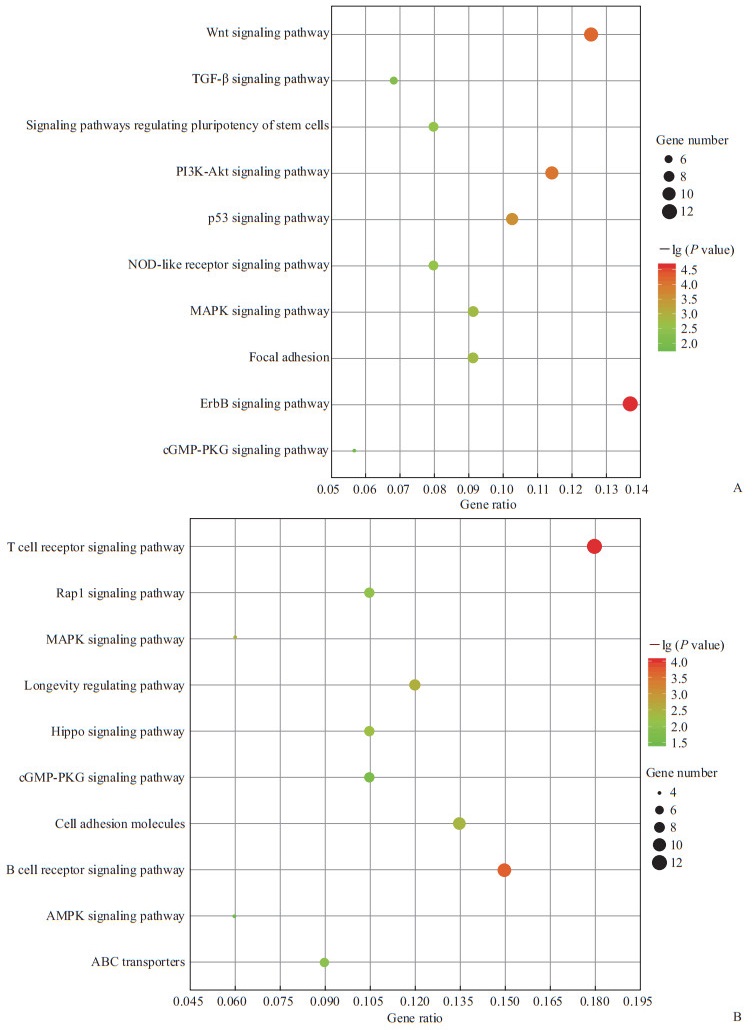
2.5 TNM分期Ⅲ+Ⅳ期与Ⅰ+Ⅱ期差异ATAC-seq数据peaks注释基因的KEGG通路富集分析结果筛选得到TNM分期Ⅲ+Ⅳ期与Ⅰ+Ⅱ期差异ATAC-seq数据peaks注释基因共384个,其中上调218个、下调166个。上调peaks注释基因显著富集的KEGG通路包括ErbB信号通路、Wnt信号通路、PI3K-Akt信号通路、P53信号通路等(图 5A),说明这些信号通路在结肠癌进展过程中持续加强,促进癌症发展。下调peaks注释基因显著富集的KEGG通路包括T细胞受体信号通路、B细胞受体信号通路、细胞黏附分子信号通路等(图 5B),这些信号通路的减弱可能与肿瘤细胞的免疫逃逸、局部扩散转移、远处脏器转移密切相关。有的信号通路在上调和下调peaks注释基因显著富集KEGG通路中均出现,如MAPK信号通路、环磷酸鸟苷-蛋白激酶G(cyclic guanosine monophosphate/protein kinase G, cGMP-PKG)信号通路等,提示其作用方式和作用效果可能有所改变。
|
图 5 结肠癌TNM分期Ⅲ+Ⅳ期相对Ⅰ+Ⅱ期上调(A)和下调(B)ATAC-seq数据peaks注释基因的KEGG通路富集分析 Fig 5 KEGG pathway enrichment analysis of up- (A) and down-regulated (B) annotated genes of colon cancer ATAC-seq peaks at TNM stage Ⅲ+Ⅳ versus stage Ⅰ+Ⅱ ATAC-seq: Assay for transposase-accessible chromatin using sequencing; KEGG: Kyoto Encyclopedia of Genes and Genomes; TGF: Transforming growth factor; PI3K-Akt: Phosphatidylinositol 3-kinase/protein kinase B; NOD: Nucleotide-binding oligomerization domain; MAPK: Mitogen-activated protein kinase; ErbB: Epidermal growth factor receptor; cGMP-PKG: Cyclic guanosine monophosphate/protein kinase G; AMPK: Adenosine monophosphate-activated protein kinase; ABC: Adenosine triphosphate-binding cassette. |
3 讨论
结肠癌作为严重威胁我国人民生命健康的最常见的消化道恶性肿瘤之一,其进展调控机制一直以来都是肿瘤科研工作者研究的热点。本研究以染色质开放状态为切入点,利用公共数据库TCGA挖掘结肠癌进展过程中染色质开放状态的变化及相关功能、信号通路的改变,研究结果对后续科研工作者研究结肠癌进展机制及治疗靶点具有十分重要的参考意义。
染色质的开放状态直接影响染色质的可及性,进而影响肿瘤的发生、发展及大量的生物学行为。Denny等[14]发现核转录因子I/B(nuclear factor I/B, Nfib)通过广泛增加的染色质可及性促进小鼠模型小细胞肺癌转移;Gomez等[15]发现干细胞能够分化出与癌症发生密切相关的染色质可及性特征,这将干细胞与人类癌症联系起来;Britton等[16]通过染色质开放状态分析证实激活蛋白1(activating protein 1, AP1)是食管腺癌的转录调节因子,在食管腺癌基因表达中发挥尤为重要的作用;Young和Trowbridge[17]则提出染色质开放状态分析可以作为鉴定癌症起源细胞的新策略。
本研究对TCGA数据库的ATAC-seq数据进行质量控制,发现测序数据peaks在染色体分布较为均匀;随后对peaks的位置特征进行归纳分析,发现位于启动子区域的peaks最多,并且大部分peaks具有多种位置特征,间接解释了转录调控的多样性。染色质开放区peaks大多位于转录起始位点附近,符合染色质开放性的特性,证实了测序数据的可靠性与真实性。
对全部结肠癌样本的ATAC-seq数据peaks进行相应位置的基因注释,并进行GO功能和KEGG通路富集分析,结果显示大量的癌症相关GO功能和KEGG信号通路出现显著富集,如Wnt信号通路的细胞间信号转导、细胞黏附分子结合相关功能、ErbB信号通路、MAPK信号通路等,反映了结肠癌发生、发展过程中的分子功能和信号通路逐渐紊乱失调,增殖信号通路呈现异常活跃状态。对目前所知的一些结肠癌关键调控基因如APC[11]、KRAS[12]、BRAF[13]启动子区的ATAC-seq数据peaks与其RNA-seq的FPKM数据进行相关性分析,结果显示启动子区染色质开放状态的peaks与其RNA-seq的FPKM呈正相关,进一步证明染色质可及性对于肿瘤的转录调控具有不可或缺的作用。
为了更进一步探索染色质开放状态在结肠癌进展过程中的变化与规律,本研究筛选了TNM分期Ⅲ+Ⅳ期相对Ⅰ+Ⅱ期有差异的染色质开放状态peaks,发现Ⅲ+Ⅳ期上调的peaks注释基因显著富集于与癌症增殖、侵袭、转移相关的信号通路,而下调的peaks注释基因则显著富集于与免疫识别相关的信号通路。这一结果提示在结肠癌进展过程中,促增殖、侵袭和转移的信号通路持续活化,而有助于对肿瘤细胞进行免疫清除的信号通路处于持续抑制、失活状态,揭示了染色质开放状态对于肿瘤生物学行为的重要影响。
综上所述,本研究利用TCGA数据库挖掘染色质开放状态与结肠癌相关基因表达调控的关系,并进一步分析了中晚期和早中期结肠癌染色质开放区的差异表达基因,从染色质开放区基因群富集的功能和信号通路角度分析了结肠癌进展过程中染色质可及性的变化,结果发现染色质开放状态对结肠癌相关功能通路起着重要作用,并有可能作为结肠癌的诊治靶标应用于临床实践,这为深入研究结肠癌调控机制提供了借鉴和参考。
| [1] |
CHEN W, ZHENG R, BAADE P D, ZHANG S, ZENG H, BRAY F, et al. Cancer statistics in China, 2015[J]. CA Cancer J Clin, 2016, 66: 115-132. DOI:10.3322/caac.21338 |
| [2] |
BRAY F, FERLAY J, SOERJOMATARAM I, SIEGEL R L, TORRE L A, JEMAL A. Global cancer statistics 2018:GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018, 68: 394-424. DOI:10.3322/caac.21492 |
| [3] |
BÜTTNER J, JÖHRENS K, KLAUSCHEN F, HUMMEL M, LENZE D, SAEGER W, et al. Intratumoral morphological heterogeneity can be an indicator of genetic heterogeneity in colorectal cancer[J]. Exp Mol Pathol, 2018, 104: 76-81. DOI:10.1016/j.yexmp.2018.01.007 |
| [4] |
PUNT C J, KOOPMAN M, VERMEULEN L. From tumour heterogeneity to advances in precision treatment of colorectal cancer[J]. Nat Rev Clin Oncol, 2017, 14: 235-246. DOI:10.1038/nrclinonc.2016.171 |
| [5] |
MOLINARI C, MARISI G, PASSARDI A, MATTEUCCI L, DE MAIO G, ULIVI P. Heterogeneity in colorectal cancer: a challenge for personalized medicine?[J/OL]. Int J Mol Sci, 2018, 19: 3733. DOI: 10.3390/ijms19123733.
|
| [6] |
JENUWEIN T, FORRESTER W C, FERNÁNDEZ-HERRERO L A, LAIBLE G, DULL M, GROSSCHEDL R. Extension of chromatin accessibility by nuclear matrix attachment regions[J]. Nature, 1997, 385: 269-272. DOI:10.1038/385269a0 |
| [7] |
PIQUE-REGI R, DEGNER J F, PAI A A, GAFFNEY D J, GILAD Y, PRITCHARD J K. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data[J]. Genome Res, 2011, 21: 447-455. DOI:10.1101/gr.112623.110 |
| [8] |
BUENROSTRO J D, GIRESI P G, ZABA L C, CHANG H Y, GREENLEAF W J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position[J]. Nat Methods, 2013, 10: 1213-1218. DOI:10.1038/nmeth.2688 |
| [9] |
BUENROSTRO J D, WU B, CHANG H Y, GREENLEAF W J. ATAC-seq: a method for assaying chromatin accessibility genome-wide[J/OL]. Curr Protoc Mol Biol, 2015, 109: 21.29.1-21.29.9. DOI: 10.1002/0471142727.mb2129s109.
|
| [10] |
TOMCZAK K, CZERWIŃSKA P, WIZNEROWICZ M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge[J/OL]. Contemp Oncol (Pozn), 2015, 19(1A): A68-A77. DOI: 10.5114/wo.2014.47136.
|
| [11] |
ZHANG L, SHAY J W. Multiple roles of APC and its therapeutic implications in colorectal cancer[J/OL]. J Natl Cancer Inst, 2017, 109: djw332. DOI: 10.1093/jnci/djw332.
|
| [12] |
BOUTIN A T, LIAO W T, WANG M, HWANG S S, KARPINETS T V, CHEUNG H, et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer[J]. Genes Dev, 2017, 31: 370-382. DOI:10.1101/gad.293449.116 |
| [13] |
RAD R, CADIÑANOS J, RAD L, VARELA I, STRONG A, KRIEGL L, et al. A genetic progression model of Braf(V600E)-induced intestinal tumorigenesis reveals targets for therapeutic intervention[J]. Cancer Cell, 2013, 24: 15-29. DOI:10.1016/j.ccr.2013.05.014 |
| [14] |
DENNY S K, YANG D, CHUANG C H, BRADY J J, LIM J S, GRÜNER B M, et al. Nfib promotes metastasis through a widespread increase in chromatin accessibility[J]. Cell, 2016, 166: 328-342. DOI:10.1016/j.cell.2016.05.052 |
| [15] |
GOMEZ N C, HEPPERLA A J, DUMITRU R, SIMON J M, FANG F, DAVIS I J. Widespread chromatin accessibility at repetitive elements links stem cells with human cancer[J]. Cell Rep, 2016, 17: 1607-1620. DOI:10.1016/j.celrep.2016.10.011 |
| [16] |
BRITTON E, ROGERSON C, MEHTA S, LI Y, LI X, OCCAMS consortium, et al. Open chromatin profiling identifies AP1 as a transcriptional regulator in oesophageal adenocarcinoma[J/OL]. PLoS Genet, 2017, 13: e1006879. DOI: 10.1371/journal.pgen.1006879.
|
| [17] |
YOUNG K, TROWBRIDGE J J. Open chromatin profiling as a novel strategy for identifying cancer cell of origin[J/OL]. Mol Cell Oncol, 2016, 3: e1236770. DOI: 10.1080/23723556.2016.1236770.
|
 2021, Vol. 42
2021, Vol. 42


