2. 海军军医大学(第二军医大学)长征医院泌尿外科, 上海 200003;
3. 海军军医大学(第二军医大学)东方肝胆外科医院泌尿外科, 上海 201805
2. Department of Urology, Changzheng Hospital, Naval Medical University (Second Military Medical University), Shanghai 200003, China;
3. Department of Urology, Eastern Hepatobiliary Surgery Hospital, Naval Medical University (Second Military Medical University), Shanghai 201805, China
肾癌是泌尿系统常见的恶性肿瘤之一,在成年人中其发病率为2%~3%,近10年来肾癌发病率和死亡率不断升高[1]。大多数肾癌发病隐匿,常因健康体检发现肾脏肿块经复查后确诊,约17%的肾癌患者在确诊时即发生转移[2]。肾癌对放射和化学治疗不敏感,治疗手段主要为手术和药物治疗,患者一旦出现肿瘤复发或耐药等情况,预后较差[3]。肿瘤干细胞指肿瘤团块中具有自我更新能力、可通过不对称分裂产生异质性肿瘤细胞亚群的一类特殊肿瘤细胞,多处于静息状态,参与肿瘤的发生、转移及耐药[4]。CD133、CD24、铜转运蛋白2(copper transport protein 2,CTR2)是肾癌干细胞的表面标志物,CD133+/CD24+/CTR2+肾癌干细胞参与了肾癌的多种生物学活动[5]。随着测序技术的发展及单细胞测序的兴起,肿瘤研究逐步从组织水平转向细胞及分子水平,基于细胞水平的预后模型能更精准地指导临床诊疗[6]。本研究通过生物信息学方法筛选肾癌干细胞相关基因,对干性基因进行功能注释、通路分析及蛋白质-蛋白质相互作用(protein-protein interaction,PPI)网络构建,发现趋化因子蛋白家族为蛋白网络的核心模块,同时进行单因素及多因素分析,确定C-X3-C基序趋化因子配体1(C-X3-C motif chemokine ligand 1,CX3CL1)可作为肾癌患者预后的独立保护因素,最后基于该分子构建肾癌预后模型。
1 资料和方法 1.1 数据获取与预处理芯片数据来源于美国Illumina公司的HumanHT-12 V4.0平台。肾透明细胞癌患者的临床信息及转录组数据均从癌症基因组图谱(The Cancer Genome Atlas,TCGA)数据库下载,TCGA中肾癌转录组数据均为标准数值矩阵,在排除预后信息缺失的患者后得到537例肾癌患者的基因表达矩阵及临床信息[7]。从基因表达汇编(Gene Expression Omnibus,GEO)数据库GSE48550数据集下载肾癌芯片数据(包含12个正常肾小管上皮细胞样本和6个CD133+/CD24+/CTR2+的肾癌干细胞样本,https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48550),使用R 3.6.3软件limma包对数据进行背景校正,数据经过以2为底的对数转化后用于后续分析。
1.2 差异表达基因分析使用R 3.6.3软件limma包分析肾癌干细胞和肾小管上皮细胞之间的差异表达基因。以校正P<0.05 [P值采用错误发现率(false discovery rate,FDR)进行校正]且|log2FC|>0.8 [FC为差异倍数(fold change)]为差异表达基因的筛选标准。
1.3 基因功能和通路富集分析使用R 3.6.3软件clusterProfiler包对差异基因进行基因本体(Gene Ontology,GO)富集分析及基因集富集分析(Gene Set Enrichment Analysis,GSEA)[8]。
1.4 PPI网络构建利用在线数据库STRING(http://string-db.org/)分析差异表达基因的PPI网络,设置最低关联度为0.8,获取关联度较高的差异表达基因。将结果导入Cytoscape软件,利用其中的cytoHubba插件确定核心蛋白网络[9]。
1.5 预后模型构建以TCGA数据库下载的肾癌患者年龄、临床分期及相关基因的表达水平为自变量,总生存情况为因变量,通过单因素及多因素Cox回归分析筛选肾癌预后的独立危险因素,并使用R 3.6.3软件rms包绘制列线图。使用Bootstrap法对列线图的预测效能进行检验。
1.6 基因差异表达验证及临床预后关联分析利用人类蛋白质图谱(Human Protein Atlas,HPA)数据库验证CX3CL1在癌组织及癌旁组织中的表达差异。使用TCGA数据库中的预后信息和基因表达情况,利用R 3.6.3软件survival包和survminer包对CX3CL1高表达和低表达(以中位数为界)的患者进行生存分析。
1.7 统计学处理采用SPSS 22.0软件进行统计学分析。通过R 3.6.3软件rms包计算C指数,评价模型对肾癌患者总体生存率的区分度。生存分析采用Kaplan-Meier法和log-rank检验。检验水准(α)为0.05。
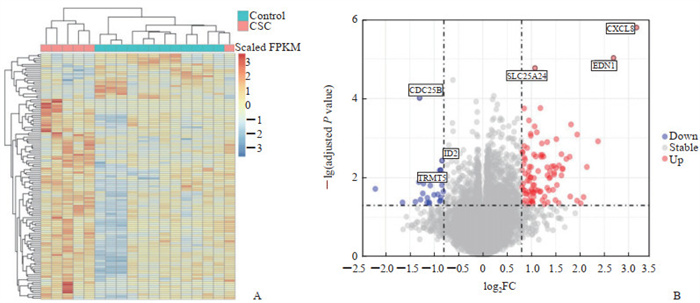
2 结果 2.1 肾癌干细胞差异表达基因分析结果通过阈值(校正P<0.05且|log2FC|>0.8)共筛选出125个差异表达基因,包括102个上调基因和23个下调基因,绘制热图及火山图,并对差异最显著的前6个基因进行了标注(图 1)。
|
图 1 基于GEO数据库GSE48550数据集的肾癌CSC差异表达基因热图及火山图 Fig 1 Heatmap and volcano map of differentially expressed genes of renal CSCs based on GSE48550 dataset of GEO database A: Heatmap of differentially expressed genes; B: Volcano map of differentially expressed genes (top 6 differentially expressed genes are marked). GEO: Gene Expression Omnibus; CSC: Cancer stem cell; FPKM: Reads per kilobase of exon model per million mapped reads; FC: Fold change; CDC25B: Cell division cycle 25B; ID2: Inhibitors of DNA binding/differentiation 2; TRMT5: Transfer RNA methyltransferase 5; SLC25A24: Solute carrier family 25 member 24; EDN1: Endothelin 1; CXCL8: C-X-C motif chemokine ligand 8. |
2.2 肾癌干细胞差异表达基因功能富集分析
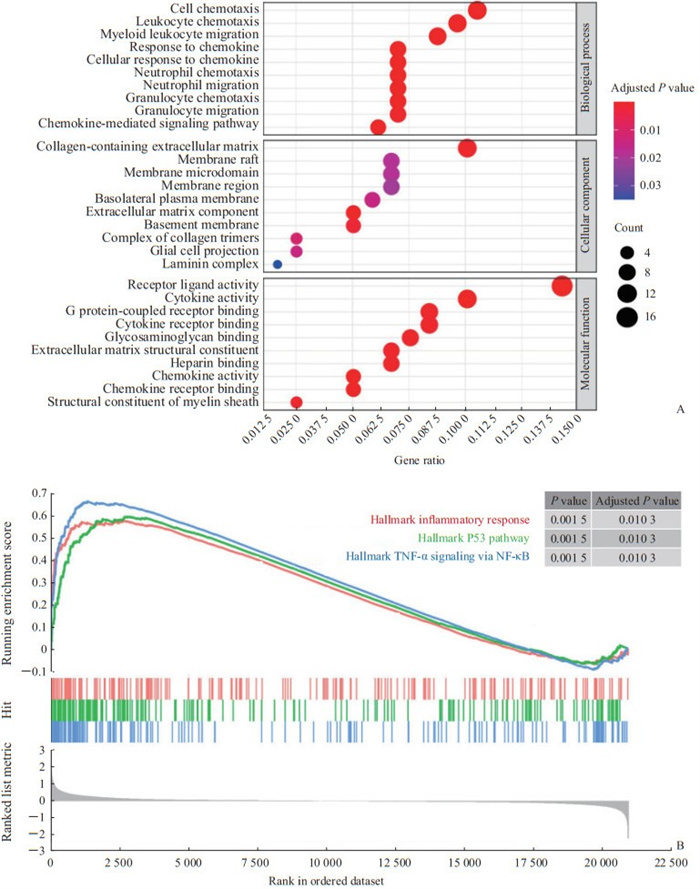
GO富集分析结果(图 2A)显示,生物过程(biological process,BP)、细胞组分(cellular component,CC)及分子功能(molecular function,MF)分别富集到2 959、244和341个条目,集中在细胞趋化、细胞外基质形成及受体配体活性等模块。GSEA分析结果(图 2B)显示,炎症反应通路、肿瘤蛋白P53通路及TNF-α/NF-κB通路在肾癌干细胞中显著激活。
|
图 2 肾癌干细胞差异表达基因的GO富集分析(A)及GSEA(B)结果 Fig 2 GO enrichment analysis (A) and GSEA (B) of differentially expressed genes of renal cancer stem cells GO: Gene Ontology; GSEA: Gene Set Enrichment Analysis; TNF-α: Tumor necrosis factor α; NF-κB: Nuclear factor κB. |
2.3 PPI网络分析
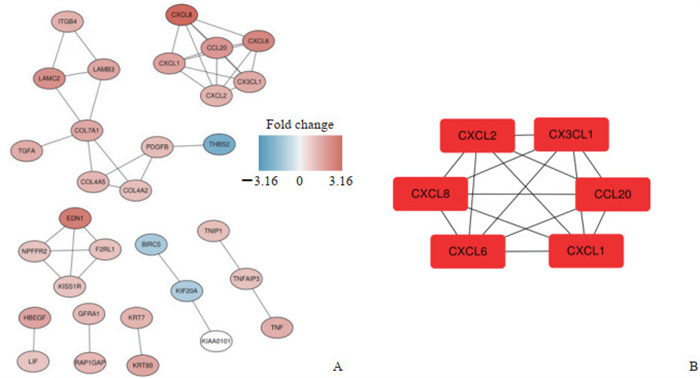
将筛选后蛋白质导入Cytoscape软件中进行重注释,得到的PPI网络图如图 3A所示。使用cytoHubba插件对PPI网络进行子网络提取,得到的核心模型如图 3B所示,可见趋化因子家族蛋白C-X-C基序趋化因子配体(C-X-C motif chemokine ligand,CXCL)2、CX3CL1、CXCL8、C-C基序趋化因子配体(C-C motif chemokine ligand,CCL)20、CXCL6、CXCL1在肾癌干细胞中富集。
|
图 3 蛋白质- 蛋白质相互作用网络图(A)及核心蛋白模块(B) Fig 3 Protein-protein interaction network diagram (A) and hub protein module (B) CXCL: C-X-C motif chemokine ligand; CX3CL1: C-X3-C motif chemokine ligand 1; CCL20: C-C motif chemokine ligand 20. |
2.4 肾癌预后的列线图模型
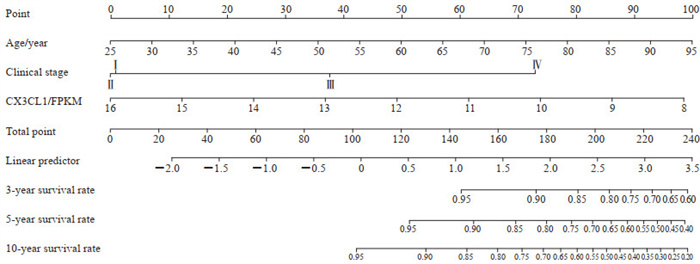
以肾癌患者年龄、临床分期及趋化因子家族蛋白为自变量,以患者生存情况为因变量进行单因素及多因素Cox回归分析,发现年龄及临床分期为肾癌预后的独立危险因素,趋化因子家族蛋白中的CX3CL1是肾癌预后的独立保护因素(表 1)。基于上述独立危险及保护因素,通过R 3.6.3软件rms包构建预测肾癌3、5及10年总体生存率的列线图(图 4),并使用Bootstrap法对列线图的预测效能进行检验,结果提示列线图对肾癌患者总体生存率的评估效能较好,C指数为0.803。
|
|
表 1 肾癌患者预后影响因素的单因素和多因素Cox回归分析 Tab 1 Univariate and multivariate Cox regression analyses of influencing factors of prognosis of patients with renal cancer |
|
图 4 预测肾癌患者3、5、10年总体生存率的列线图模型 Fig 4 Nomogram model for predicting 3-, 5- and 10-year overall survival rates of renal cancer patients CX3CL1: C-X3-C motif chemokine ligand 1; FPKM: Reads per kilobase of exon model per million mapped reads. |
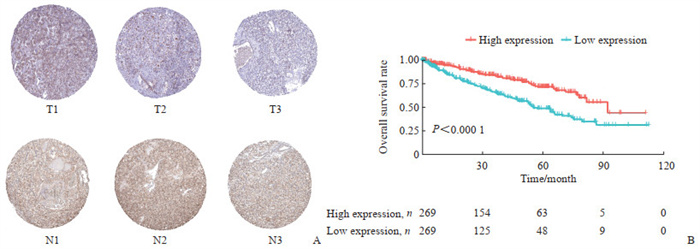
2.5 肾癌患者CX3CL1蛋白表达水平验证及其与预后的关系
通过HPA数据库发现CX3CL1在肾癌中的表达量低于正常肾脏组织(图 5A)。根据CX3CL1的表达水平将患者分为低表达组和高表达组,生存分析结果显示,CX3CL1高表达组患者总生存期长于低表达组(P<0.000 1,图 5B)。
|
图 5 肾癌组织中CX3CL1的表达及其与患者生存的关系 Fig 5 Expression of CX3CL1 in renal cancer tissues and its relationship with patient survival A: CX3CL1 expression in renal cancer and normal renal tissues. T1-T3 represent renal cancer tissues, and N1-N3 represent normal renal tissues. B: Overall survival analysis of renal cancer patients with high or low expression of CX3CL1. CX3CL1: C-X3-C motif chemokine ligand 1. |
3 讨论
肾癌是一种异质性显著、发病机制复杂的恶性肿瘤,多种基因参与了肾癌的发病,生物信息学的发展及大型测序队列数据的公布为肾癌发病机制研究及模型构建提供了基础。肿瘤干细胞具有器官特异性,通过损伤修复、代谢调整、突变累积等机制在肿瘤进程中发挥作用。采用单细胞测序技术结合时序分析鉴定出了大量的肿瘤起始细胞,后者是肿瘤诊疗的新靶标。本研究通过GEO及TCGA两大数据库筛选肾癌干细胞相关基因,发现CX3CL1表达是肾癌预后的独立保护因素,并将该蛋白与其他临床指标结合构建了一种新的肾癌预后模型。
CX3CL1属于趋化因子蛋白家族成员,该蛋白家族参与了肿瘤细胞增殖、迁移和侵袭。Conley-Lacomb等[10-11]发现CXCL12/C-X-C基序趋化因子受体(C-X-C motif chemokine receptor,CXCR)4轴促进前列腺癌转移;CCL2作用于血管内皮细胞,通过Janus激酶2(Janus kinase 2,JAK2)-信号转导与转录激活因子5(signal transducer and activator of transcription 5,STAT5)和MAPK通路促进肿瘤的血管生成和转移。Zhu等[12]和Biswas等[13]发现CXCL13/CXCR5轴参与乳腺癌、结肠癌和淋巴瘤的进展。趋化因子蛋白可作用于免疫系统,影响肿瘤进程。Qian等[14]发现CCL2通过招募炎性单核细胞促进乳腺癌转移。研究发现肿瘤相关巨噬细胞通过分泌CCL18促进卵巢癌、乳腺癌和胰腺导管腺癌转移[15-16]。Gao等[17]发现CXCL5作用于CXCR5促进膀胱癌细胞分泌基质金属蛋白酶(matrix metalloproteinase,MMP)2和MMP9,后者促进肿瘤转移和侵袭。Maxwell等[18]发现CXCL8可促进前列腺癌细胞在缺氧环境中存活。CX3CL1蛋白通过转录后修饰形成多种变体,均通过结合C-X3-C基序趋化因子受体1(C-X3-C motif chemokine receptor 1,CX3CR1)发挥生物学功能[19]。CX3CL1是一种跨膜糖蛋白,在人体中以膜结合型和可溶型2种方式存在,其中膜结合型CX3CL1可发挥黏附功能,促进内皮细胞与CX3CR1+细胞黏附;可溶型CX3CL1是膜结合型被去整合素和金属蛋白酶(a disintegrin and metalloproteinase,ADAM)10和ADAM17酶切而成,介导白细胞趋化等[20]。目前对CX3CL1在肿瘤中的具体机制观点不一,有研究发现CX3CL1发挥抗肿瘤作用,也有研究证实CX3CL1促进肿瘤进展[21]。本研究发现该分子是肾癌患者预后的保护因素。
本研究也存在一些局限性:首先,缺乏CX3CL1在肾癌中的机制研究;其次,没有考虑CX3CL1在其他组学,如体细胞突变、结构变异及甲基化修饰方面的变异;最后,本研究没有在中国肾癌队列中验证CX3CL1是否为肾癌患者的预后保护因素。上述分析将在后续研究中完成。总而言之,本研究通过数据挖掘发现CX3CL是肾癌患者预后的独立保护因素,为肾癌干细胞研究提供了新的方向。
| [1] |
SIEGEL R L, MILLER K D, JEMAL A. Cancer statistics, 2019[J]. CA Cancer J Clin, 2019, 69: 7-34. DOI:10.3322/caac.21551 |
| [2] |
MASON R J, WOOD L, KAPOOR A, BASAPPA N, BJARNASON G, BOORJIAN S A, et al. Kidney Cancer Research Network of Canada (KCRNC) consensus statement on the role of cytoreductive nephrectomy for patients with metastatic renal cell carcinoma[J]. Can Urol Assoc J, 2019, 13: 166-174. |
| [3] |
BALDEWIJNS M M, VAN VLODROP I J, SCHOUTEN L J, SOETEKOUW P M, DE BRUÏNE A P, VAN ENGELAND M. Genetics and epigenetics of renal cell cancer[J]. Biochim Biophys Acta, 2008, 1785: 133-155. |
| [4] |
VLASHI E, PAJONK F. Cancer stem cells, cancer cell plasticity and radiation therapy[J]. Semin Cancer Biol, 2015, 31: 28-35. DOI:10.1016/j.semcancer.2014.07.001 |
| [5] |
岳中瑾. 肿瘤干细胞及肾癌相关研究进展[J]. 中华泌尿外科杂志, 2005, 26: 573-575. DOI:10.3760/j:issn:1000-6702.2005.08.022 |
| [6] |
XIAO W, GAO Z Y, DUAN Y X, YUAN W X, KE Y. Notch signaling plays a crucial role in cancer stem-like cells maintaining stemness and mediating chemotaxis in renal cell carcinoma[J/OL]. J Exp Clin Cancer Res, 2017, 36: 41. DOI: 10.1186/s13046-017-0507-3.
|
| [7] |
RITCHIE M E, PHIPSON B, WU D, HU Y, LAW C W, SHI W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies[J/OL]. Nucleic Acids Res, 2015, 43: e47. DOI: 10.1093/nar/gkv007.
|
| [8] |
YU G, WANG L G, HAN Y, HE Q Y. clusterProfiler: an R package for comparing biological themes among gene clusters[J]. OMICS, 2012, 16: 284-287. DOI:10.1089/omi.2011.0118 |
| [9] |
SHANNON P, MARKIEL A, OZIER O, BALIGA N S, WANG J T, RAMAGE D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks[J]. Genome Res, 2003, 13: 2498-2504. DOI:10.1101/gr.1239303 |
| [10] |
CONLEY-LACOMB M K, SEMAAN L, SINGAREDDY R, LI Y F, HEATH E I, KIM S, et al. Pharmacological targeting of CXCL12/CXCR4 signaling in prostate cancer bone metastasis[J/OL]. Mol Cancer, 2016, 15: 68. DOI: 10.1186/s12943-016-0552-0.
|
| [11] |
CONLEY-LACOMB M K, SALIGANAN A, KANDAGATLA P, CHEN Y Q, CHER M L, CHINNI S R. PTEN loss mediated Akt activation promotes prostate tumor growth and metastasis via CXCL12/CXCR4 signaling[J/OL]. Mol Cancer, 2013, 12: 85. DOI: 10.1186/1476-4598-12-85.
|
| [12] |
ZHU Z Y, ZHANG X K, GUO H L, FU L, PAN G L, SUN Y G. CXCL13-CXCR5 axis promotes the growth and invasion of colon cancer cells via PI3K/AKT pathway[J]. Mol Cell Biochem, 2015, 400: 287-295. DOI:10.1007/s11010-014-2285-y |
| [13] |
BISWAS S, SENGUPTA S, ROY CHOWDHURY S, JANA S, MANDAL G, MANDAL P K, et al. CXCL13-CXCR5 co-expression regulates epithelial to mesenchymal transition of breast cancer cells during lymph node metastasis[J]. Breast Cancer Res Treat, 2014, 143: 265-276. DOI:10.1007/s10549-013-2811-8 |
| [14] |
QIAN B Z, LI J F, ZHANG H, KITAMURA T, ZHANG J H, CAMPION L R, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis[J]. Nature, 2011, 475: 222-225. DOI:10.1038/nature10138 |
| [15] |
CHEN J, YAO Y, GONG C, YU F, SU S, CHEN J, et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3[J]. Cancer Cell, 2011, 19: 541-555. DOI:10.1016/j.ccr.2011.02.006 |
| [16] |
XIAO Y C, YANG Z B, CHENG X S, FANG X B, SHEN T, XIA C F, et al. CXCL8, overexpressed in colorectal cancer, enhances the resistance of colorectal cancer cells to anoikis[J]. Cancer Lett, 2015, 361: 22-32. DOI:10.1016/j.canlet.2015.02.021 |
| [17] |
GAO Y, GUAN Z F, CHEN J Q, XIE H J, YANG Z, FAN J H, et al. CXCL5/CXCR2 axis promotes bladder cancer cell migration and invasion by activating PI3K/AKT-induced upregulation of MMP2/MMP9[J]. Int J Oncol, 2015, 47: 690-700. DOI:10.3892/ijo.2015.3041 |
| [18] |
MAXWELL P J, COULTER J, WALKER S M, MCKECHNIE M, NEISEN J, MCCABE N, et al. Potentiation of inflammatory CXCL8 signalling sustains cell survival in PTEN-deficient prostate carcinoma[J]. Eur Urol, 2013, 64: 177-188. DOI:10.1016/j.eururo.2012.08.032 |
| [19] |
SCHMALL A, AL-TAMARI H M, HEROLD S, KAMPSCHULTE M, WEIGERT A, WIETELMANN A, et al. Macrophage and cancer cell cross-talk via CCR2 and CX3CR1 is a fundamental mechanism driving lung cancer[J]. Am J Respir Crit Care Med, 2015, 191: 437-447. DOI:10.1164/rccm.201406-1137OC |
| [20] |
GUNNER G, CHEADLE L, JOHNSON K M, AYATA P, BADIMON A, MONDO E, et al. Sensory lesioning induces microglial synapse elimination via ADAM10 and fractalkine signaling[J]. Nat Neurosci, 2019, 22: 1075-1088. DOI:10.1038/s41593-019-0419-y |
| [21] |
CONROY M J, LYSAGHT J. CX3CL1 signaling in the tumor microenvironment[J]. Adv Exp Med Biol, 2020, 1231: 1-12. |
 2021, Vol. 42
2021, Vol. 42


