2. 重庆大学附属肿瘤医院/肿瘤研究所肿瘤转移与个体化诊治转化研究重庆市重点实验室, 重庆 400030
2. Chongqing Key Laboratory of Translational Research for Cancer Metastasis and Individualized Treatment, Chongqing University Cancer Hospital/Chongqing Cancer Research Institute, Chongqing 400030, China
乳腺癌是女性最常见的恶性肿瘤,在我国的发病率占所有新发癌症病例的15%,且死亡率呈逐年上升趋势,严重危害女性健康[1-2]。目前用于乳腺癌筛查的肿瘤标志物主要是癌抗原15-3(cancer antigen 15-3,CA15-3),但其对乳腺癌早期筛查缺乏特异性,往往在中、晚期乳腺癌患者中呈高表达[3]。因此,寻找乳腺癌筛查相对特异的标志物具有重要意义。细胞功能的病理改变常伴有代谢重组,包括氨基酸代谢的改变[4]。氨基酸检测对许多遗传性代谢性疾病的筛查非常重要。癌症患者处于高代谢状态和高分解代谢状态,蛋白的合成和分解较健康人群均有所提高,从而导致氨基酸浓度的变化和氨基酸代谢异常[5]。研究表明,氨基酸在筛查肿瘤方面是一个非常有潜力的生物标志物,在肺癌[6-7]、卵巢癌[8]、胃癌[9]、结直肠癌[10]、胰腺癌[11]等肿瘤中已有报道,但在乳腺癌筛查中的应用国内尚缺乏系统性研究。为探讨血清氨基酸在乳腺癌患者与正常人之间水平的差异及其在乳腺癌筛查中的价值,本实验用高效液相色谱-质谱法(high performance liquid chromatography-mass spectrometry,HPLC-MS)对乳腺癌患者和正常人血清中20种氨基酸水平进行检测,分析氨基酸水平的差异,并对差异血清氨基酸在乳腺癌筛查中的效能进行系统性评估,以寻找新的乳腺癌筛查生物标志物。
1 资料和方法 1.1 一般资料招募2019年4-6月经重庆大学附属肿瘤医院病理科确诊且未经任何治疗的乳腺癌患者59例作为乳腺癌组,均为女性浸润性导管癌患者,年龄为44~64岁(中位年龄51岁)。根据美国癌症联合委员会(American Joint Committee on Cancer,AJCC)癌症分期手册第8版对患者进行TNM分期,59例中包括早期乳腺癌患者26例(TNM分期Ⅰ期6例、Ⅱ期20例)、晚期乳腺癌患者33例(TNM分期Ⅲ期23例、Ⅳ期10例)。收集患者临床病理资料并对其进行分子分型,其中Luminal A型16例、Luminal B型25例、三阴性型8例、人表皮生长因子受体2(human epidermal growth factor receptor 2,HER2)阳性型10例。同时招募同期重庆大学附属肿瘤医院健康体检中心正常女性体检者53例作为对照组,年龄为36~62岁(中位年龄47岁)。本研究通过重庆大学附属肿瘤医院伦理委员会审批,所有研究方案均得到招募者同意并签订知情同意书。
1.2 样本采集于清晨空腹时,采集招募者前臂肘正中静脉血3 mL于未加任何抗凝剂的真空采血管中,室温静置0.5 h,待血液凝固后,1 200×g离心10 min分离血清(所有样本离心分离均在样本采集2 h内完成),取300 μL上清液于-80 ℃低温冰箱中冻存待用。
1.3 主要试剂及仪器Thermo Ultimate 3000液相色谱仪串联Thermo TSQ Endura质谱仪(美国ThermoFisher Scientific公司);HPLC-MS氨基酸标准品和内标(美国剑桥同位素实验室)。
1.4 检测指标对血清中20种氨基酸进行检测,包括8种必需氨基酸,即赖氨酸(lysine,Lys)、色氨酸(tryptophan,Trp)、苯丙氨酸(phenylalanine,Phe)、甲硫氨酸(methionine,Met)、苏氨酸(threonine,Thr)、异亮氨酸(isoleucine,Ile)、亮氨酸(leucine,Leu)、缬氨酸(valine,Val);12种非必需氨基酸,即精氨酸(arginine,Arg)、组氨酸(histidine,His)、丝氨酸(serine,Ser)、酪氨酸(tyrosine,Tyr)、谷氨酸(glutamic acid,Glu)、鸟氨酸(ornithine,Orn)、丙氨酸(alanine,Ala)、脯氨酸(proline,Pro)、瓜氨酸(citrulline,Cit)、天冬酰胺(asparagine,Asn)、谷氨酰胺(glutamine,Gln)、天冬氨酸(aspartic acid,Asp)。
将沉淀剂与内标储备液按照13∶1的比例混合均匀,另取大于25 mL的超纯水,备用。样本准备:取出V型96孔深孔板,往相应孔中分别加入40 μL校准品/空白样本/质控样本(双空白即为180 μL超纯水;空白即为40 μL超纯水+140 μL沉淀剂);沉淀:用多道移液器向96孔板中加入140 μL沉淀剂,封膜涡旋30 s,重复2次,1 200×g 4℃离心30 min;稀释:取20 μL上清液至相对应的另一块96孔板中(-18 ℃可保存1 d),加入180 μL超纯水稀释,封膜涡旋30 s,重复2次后1 200×g离心3 min,上机测定。
1.5 实验仪器参数液相色谱条件:ACQUITY UPLC HSS T3 VanGuard保护柱(10 nm,1.8 μm,2.1 mm×5 mm,美国Waters公司),ACQUITY UPLC HSS T3色谱柱(2.1 mm×100 mm,1.8 μm,美国Waters公司),柱温45 ℃,流速0.4 mL/min,进样量10 μL,自动进样器温度10 ℃。流动相A液为含0.1%甲酸、0.05%七氟丁酸的ddH2O,流动相B液为含0.1%甲酸、0.05%七氟丁酸的乙腈溶液,梯度洗脱条件:0~0.5 min,98% A;0.5~1.5 min,98% A到90% A;1.5~3.5 min,90% A到65% A;3.5~3.6 min,65% A到5% A;3.6~4.8 min,5% A;4.8~5 min,5% A到98% A;5~6 min,98% A。质谱条件:离子源为电喷雾(electrospray ionization,ESI),离子源喷雾电压为3.5 kV,鞘气流速为50 arb,辅助气流速为15 arb,反向气流速为1 arb,传输毛细管温度为350 ℃,辅助气加热温度为400 ℃。扫描模式为多反应监测(multiple reaction monitoring,MRM)。
1.6 统计学处理用SPSS 23.0软件进行统计学分析。计量资料如服从正态分布采用x±s表示,正常人与乳腺癌患者和早、晚期乳腺癌患者血清氨基酸水平差异分析采用独立样本t检验;乳腺癌患者不同分子亚型血清氨基酸水平差异比较采用单因素方差分析,两两比较采用最小显著性差异法。14种血清氨基酸及其联合筛查效能采用ROC曲线进行评估,取约登指数(灵敏度与特异度之和减去1)最大的一点为最佳截断值,并计算其对应筛查灵敏度和特异度。检验水准(α)为0.05。
2 结果 2.1 对照组与乳腺癌组血清中20种氨基酸水平的比较乳腺癌组血清中Ala、Asn、Cit、Glu、His、Ile、Leu、Lys、Orn、Phe、Pro、Thr、Tyr、Val的水平均高于对照组,差异有统计学意义(P均<0.05);乳腺癌组血清中Arg的水平低于对照组,差异有统计学意义(P=0.003)。见图 1。

|
图 1 对照组与乳腺癌组血清氨基酸水平比较 Fig 1 Comparison of serum amino acid levels between control group and breast cancer group *P < 0.05, **P < 0.01. x±s. Ala: Alanine; Arg: Arginine; Asn: Asparagine; Asp: Aspartic acid; Cit: Citrulline; Gln: Glutamine; Glu: Glutamic acid; His: Histidine; Ile: Isoleucine; Leu: Leucine; Lys: Lysine; Met: Methionine; Orn: Ornithine; Phe: Phenylalanine; Pro: Proline; Ser: Serine; Thr: Threonine; Trp: Tryptophan; Tyr: Tyrosine; Val: Valine. |
2.2 血清氨基酸在乳腺癌筛查中的效能评估结果
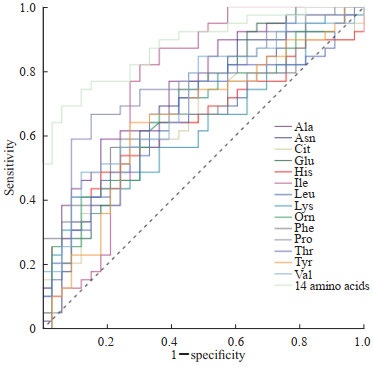
ROC曲线分析表明,在乳腺癌患者血清中升高的14种氨基酸(Ala、Asn、Cit、Glu、His、Ile、Leu、Lys、Orn、Phe、Pro、Thr、Tyr、Val)中,Ala和Pro单独筛查乳腺癌时的效能最高,AUC均为0.75;Ile单独筛查乳腺癌时灵敏度最高,达0.82;Lys单独筛查乳腺癌时特异度最高,达0.94;14种氨基酸联合检测能够提高乳腺癌患者的筛查效能,AUC为0.88,灵敏度为0.69,特异度为0.94。见图 2、表 1。
|
图 2 乳腺癌患者血清中升高的14种氨基酸单独及联合检测筛查乳腺癌的ROC曲线 Fig 2 ROC curves of 14 elevated serum amino acids alone and their combined detection for screening of breast cancer ROC: Receiver operating characteristic; Ala: Alanine; Asn: Asparagine; Cit: Citrulline; Glu: Glutamic acid; His: Histidine; Ile: Isoleucine; Leu: Leucine; Lys: Lysine; Orn: Ornithine; Phe: Phenylalanine; Pro: Proline; Thr: Threonine; Tyr: Tyrosine; Val: Valine. |
|
|
表 1 乳腺癌患者血清中升高的14种氨基酸单独及联合检测筛查乳腺癌的效能评估 Tab 1 Screening performance of 14 elevated serum amino acids alone and their combined detection for breast cancer |
2.3 早期和晚期乳腺癌患者血清中20种氨基酸水平比较
早期(TNM分期Ⅰ期和Ⅱ期)、晚期(TNM分期Ⅲ期和Ⅳ期)乳腺癌患者血清氨基酸水平比较结果表明,前者Val水平高于后者,差异有统计学意义(P=0.044);其余19种氨基酸水平在早期和晚期乳腺癌患者之间差异均无统计学意义(P均>0.05,表 2)。
|
|
表 2 早期(TNM分期Ⅰ ~Ⅱ期)和晚期(TNM分期Ⅲ ~Ⅳ期)乳腺癌患者血清中20种氨基酸水平的比较 Tab 2 Levels of 20 serum amino acids in early (TNM stage Ⅰ -Ⅱ) and advanced (TNM stage Ⅲ-Ⅳ) breast cancer patients |
2.4 不同分子亚型的乳腺癌患者血清中20种氨基酸水平比较
比较乳腺癌Luminal A型、Luminal B型、三阴性型、HER2阳性型患者血清氨基酸水平,结果显示20种氨基酸在乳腺癌各分子亚型中的水平差异均无统计学意义(P均>0.05,表 3)。
|
|
表 3 不同分子亚型乳腺癌患者血清中20种氨基酸水平比较 Tab 3 Levels of 20 serum amino acids in breast cancer patients with different molecular subtypes |
3 讨论
氨基酸是合成蛋白质的基本原材料和蛋白质代谢的产物,氨基酸代谢是调节生长、繁殖和免疫所必需的关键代谢途径[12]。肿瘤患者因肿瘤炎症刺激产生急性时相反应蛋白的增加及厌食情绪、吞咽困难、消化不良等会导致机体对蛋白和能量的需求改变,并伴随代谢紊乱[4]。代谢组学分析是一种非常有前途的用于肿瘤早期筛查的方法。越来越多的研究表明氨基酸是一类极具潜力的生物标志物,已被用于恶性肿瘤的筛查和相关发病机制的研究[4, 6-11, 13-15],但在乳腺癌相关研究中仅有少量小样本报道,尚存在争议。Poschke等[16]采用高效液相色谱法对41例乳腺癌患者和9例乳腺良性病变患者术前、术后15种血清氨基酸水平进行了测定,结果发现与乳腺良性病变患者相比,乳腺癌患者术前血清中8种氨基酸(Glu、Ser、Gln、Ala、Val、Phe、Ile、Leu)水平升高。Miyagi等[17]研究了196例乳腺癌患者和976例正常人血浆氨基酸水平,发现乳腺癌患者血浆Thr、Pro、Ser、Gly、Ala和Orn水平高于正常人,而Gln、Trp、His、Phe和Tyr水平却比正常人低。Gu等[18]检测了28例乳腺癌和137例健康对照者血浆中氨基酸水平,乳腺癌组Arg和Thr水平高于健康对照组,而Gly、Asp、His、Gln、Pro水平低于健康对照组。本实验用HPLC-MS法对乳腺癌患者和正常人血清中的20种氨基酸进行了检测,发现乳腺癌患者有14种血清氨基酸(Ala、Asn、Cit、Glu、His、Ile、Leu、Lys、Orn、Phe、Pro、Thr、Tyr、Val)水平高于正常人(P均<0.05),仅有Arg水平低于正常人(P=0.003)。各研究结果不一致的原因可能与研究人群来源、样本检测量及检测方法不同有关。但本研究及文献报道的结果均证实乳腺癌患者血液中氨基酸代谢与正常人有差异,具有用于乳腺癌筛查的潜力。
乳腺癌是女性发病率最高的肿瘤,严重危害女性健康[1-2]。高死亡率和较差的预后主要由于滞后的诊断,因此,及早地诊断乳腺癌是改善乳腺癌患者整体生存率的有效途径。目前乳腺癌健康筛查主要依靠影像学如超声、钼靶等,缺乏特异的血清学标志物。本实验对乳腺癌患者血清氨基酸中高于正常人的14种差异氨基酸单独及联合检测绘制了ROC曲线,计算筛查最佳截断值及对应的筛查灵敏度和特异度,并用AUC评估其筛查乳腺癌的效能,结果显示Ala、Pro单独筛查乳腺癌的效能较高(AUC均为0.75),Ile单独筛查乳腺癌时拥有最高灵敏度,Lys拥有最高特异度。本研究结果显示没有哪一种血清氨基酸单独筛查乳腺癌时同时拥有比较理想的灵敏度和特异度,但14种氨基酸联合检测时能够提高乳腺癌患者的筛查效能,因此建议在用血清氨基酸对乳腺癌进行筛查时采用多种氨基酸联合检测。
乳腺癌的分期直接关系到患者的生存预后,本研究对早期和晚期乳腺癌患者血清氨基酸水平进行了比较,发现前者血清氨基酸Val水平高于后者,同时Val在乳腺癌血清中整体水平高于正常人,表明血清氨基酸Val水平对于早期乳腺癌筛查具有非常大的潜力。
根据乳腺肿瘤组织中4个分子(HER2、雌激素受体、孕激素受体和Ki-67)的免疫组织化学染色结果,可将乳腺癌分为不同的分子亚型(Luminal A型、Luminal B型、三阴性型、HER2阳性型),不同分子亚型其临床治疗方案及预后均有所不同[19-20]。有研究表明血清氨基酸水平在侵袭性最强的乳腺癌亚型(基底细胞型)中较侵袭性弱的乳腺癌亚型(Luminal A型)高[16],但本研究对乳腺癌Luminal A型、Luminal B型、三阴性型、HER2阳性型乳腺癌患者血清氨基酸水平的比较结果发现,在所有分子亚型乳腺癌中20种血清氨基酸水平差异均无统计学意义(P均>0.05)。血清氨基酸在不同分子亚型乳腺癌中的表达差异还需进一步证实。
过去几十年里,尽管乳腺癌的死亡率随着新的治疗手段的发展而降低,但没有任何治疗手段对所有分子亚型的乳腺癌都有效[20]。近年来可用于多种乳腺癌分子亚型治疗的代谢饥饿疗法逐渐走进人们的视野,这种治疗手段通过去除或限制某些特殊的代谢物以达到有效治疗的目的,与传统的化学治疗和放射治疗相比不良反应较低,是一种非常有潜力的治疗策略[21]。氨基酸失衡疗法是一种非常有前景的代谢饥饿疗法,通过去除、减少或增加某些特殊氨基酸既能够选择性地抑制肿瘤细胞的生长,又能改患者营养状况,最终达到治疗肿瘤的目的[22-24]。本研究显示乳腺癌患者血清氨基酸中14种血清氨基酸水平高于正常人,说明正常人中一些非必需氨基酸可能是肿瘤生长的必需氨基酸,有意识地减少、抑制或去除这些氨基酸的摄入可能有助于乳腺癌治疗。有趣的是,本研究发现乳腺癌患者血清中非必需氨基酸Arg水平低于正常人,这与Vissers等[25]研究结果类似。然而有研究显示乳腺癌组织中氨基酸Arg水平比乳腺良性组织高,有些肿瘤中由于缺乏精氨酸琥珀酸合成酶无法合成Arg,因此Arg对某些肿瘤来说是必需氨基酸[26-28]。Arg代谢与很多其他代谢途径相互作用能够促进肿瘤的生长[29-31],出现乳腺癌组织中Arg水平高而血清中Arg水平相对较低的情况,可能由于乳腺癌的生长必需Arg参与,而乳腺癌患者体内又缺乏精氨酸琥珀酸合成酶无法合成Arg,导致血清中Arg水平降低。据此推测,通过某些治疗手段耗尽体内Arg也许对乳腺癌治疗有利,Arg可能是乳腺癌新的治疗靶点。
综上所述,血清氨基酸水平在乳腺癌与正常人中存在差异,可用于乳腺癌的筛查,多种血清氨基酸同时检测有利于提高乳腺癌的筛查效能,是非常有潜力的生物标志物。本研究的病例数相对较少,仅进行了探索性研究,血清氨基酸能否用于乳腺癌筛查的临床实践还需要后续大样本验证。
| [1] |
CHEN W, ZHENG R, BAADE P D, ZHANG S, ZENG H, BRAY F, et al. Cancer statistics in China, 2015[J]. CA Cancer J Clin, 2016, 66: 115-132. DOI:10.3322/caac.21338 |
| [2] |
ALLEMANI C, MATSUDA T, DI CARLO V, HAREWOOD R, MATZ M, NIKŠIĆ M, et al. Global surveillance of trends in cancer survival 2000-14(CONCORD-3): analysis of individual records for 37513025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries[J]. Lancet, 2018, 391: 1023-1075. DOI:10.1016/S0140-6736(17)33326-3 |
| [3] |
TANG S, WEI L, SUN Y, ZHOU F, ZHU S, YANG R, et al. CA153 in breast secretions as a potential molecular marker for diagnosing breast cancer: a meta analysis[J/OL]. PLoS One, 2016, 11: e0163030. DOI: 10.1371/journal.pone.0163030.
|
| [4] |
MANIG F, KUHNE K, VON NEUBECK C, SCHWARZENBOLZ U, YU Z R, KESSLER B M, et al. The why and how of amino acid analytics in cancer diagnostics and therapy[J]. J Biotechnol, 2017, 242: 30-54. DOI:10.1016/j.jbiotec.2016.12.001 |
| [5] |
CHENG F, WANG Z, HUANG Y, DUAN Y, WANG X. Investigation of salivary free amino acid profile for early diagnosis of breast cancer with ultra performance liquid chromatography-mass spectrometry[J]. Clin Chim Acta, 2015, 447: 23-31. DOI:10.1016/j.cca.2015.05.008 |
| [6] |
SHINGYOJI M, IIZASA T, HIGASHIYAMA M, IMAMURA F, SARUKI N, IMAIZUMI A, et al. The significance and robustness of a plasma free amino acid (PFAA) profile-based multiplex function for detecting lung cancer[J/OL]. BMC Cancer, 2013, 13: 77. DOI: 10.1186/1471-2407-13-77.
|
| [7] |
ZHAO Q H, CAO Y, WANG Y, HU C L, HU A L, RUAN L, et al. Plasma and tissue free amino acid profiles and their concentration correlation in patients with lung cancer[J]. Asia Pac J Clin Nutr, 2014, 23: 429-436. |
| [8] |
PLEWA S, HORAŁA A, DEREZIŃSKI P, KLUPCZYNSKA A, NOWAK-MARKWITZ E, MATYSIAK J, et al. Usefulness of amino acid profiling in ovarian cancer screening with special emphasis on their role in cancerogenesis[J/OL]. Int J Mol Sci, 2017, 18: 2727. DOI: 10.3390/ijms18122727.
|
| [9] |
JING F, HU X, CAO Y, XU M, WANG Y, JING Y, et al. Discriminating gastric cancer and gastric ulcer using human plasma amino acid metabolic profile[J]. IUBMB Life, 2018, 70: 553-562. DOI:10.1002/iub.1748 |
| [10] |
GAO P, ZHOU C, ZHAO L, ZHANG G, ZHANG Y. Tissue amino acid profile could be used to differentiate advanced adenoma from colorectal cancer[J]. J Pharm Biomed Anal, 2016, 118: 349-355. DOI:10.1016/j.jpba.2015.11.007 |
| [11] |
TUMAS J, BASKIROVA I, PETRENAS T, NORKUNIENE J, STRUPAS K, SILEIKIS A. Towards a personalized approach in pancreatic cancer diagnostics through plasma amino acid analysis[J]. Anticancer Res, 2019, 39: 2035-2042. DOI:10.21873/anticanres.13314 |
| [12] |
VAN DER MEIJ B S, TELENI L, ENGELEN M P K J, DEUTZ N E P. Amino acid kinetics and the response to nutrition in patients with cancer[J]. Int J Radiat Biol, 2019, 95: 480-492. DOI:10.1080/09553002.2018.1466209 |
| [13] |
SIMIŃSKA E, KOBA M. Amino acid profiling as a method of discovering biomarkers for early diagnosis of cancer[J]. Amino Acids, 2016, 48: 1339-1345. DOI:10.1007/s00726-016-2215-2 |
| [14] |
BERNFELD E, FOSTER D A. Glutamine as an essential amino acid for KRas-driven cancer cells[J]. Trends Endocrinol Metab, 2019, 30: 357-368. DOI:10.1016/j.tem.2019.03.003 |
| [15] |
SAITO Y, MORIYA S, KAZAMA H, HIRASAWA K, MIYAHARA K, KOKUBA H, et al. Amino acid starvation culture condition sensitizes EGFR-expressing cancer cell lines to gefitinib-mediated cytotoxicity by inducing atypical necroptosis[J]. Int J Oncol, 2018, 52: 1165-1177. |
| [16] |
POSCHKE I, MAO Y, KIESSLING R, DE BONIFACE J. Tumor-dependent increase of serum amino acid levels in breast cancer patients has diagnostic potential and correlates with molecular tumor subtypes[J/OL]. J Transl Med, 2013, 11: 290. DOI: 10.1186/1479-5876-11-290.
|
| [17] |
MIYAGI Y, HIGASHIYAMA M, GOCHI A, AKAIKE M, ISHIKAWA T, MIURA T, et al. Plasma free amino acid profiling of five types of cancer patients and its application for early detection[J/OL]. PLoS One, 2011, 6: e24143. DOI: 10.1371/journal.pone.0024143.
|
| [18] |
GU Y, CHEN T, FU S, SUN X, WANG L, WANG J, et al. Perioperative dynamics and significance of amino acid profiles in patients with cancer[J/OL]. J Transl Med, 2015, 13: 35. DOI: 10.1186/s12967-015-0408-1.
|
| [19] |
GOLDHIRSCH A, WINER E P, COATES A S, GELBER R D, PICCART-GEBHART M, THÜRLIMANN B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013[J]. Ann Oncol, 2013, 24: 2206-2223. DOI:10.1093/annonc/mdt303 |
| [20] |
TELLI M L, GRADISHAR W J, WARD J H. NCCN guidelines updates: breast cancer[J]. J Natl Compr Canc Netw, 2019, 17: 552-555. |
| [21] |
CHANGOU C A, CHEN Y R, XING L, YEN Y, CHUANG F Y, CHENG R H, et al. Arginine starvation-associated atypical cellular death involves mitochondrial dysfunction, nuclear DNA leakage, and chromatin autophagy[J]. Proc Natl Acad Sci U S A, 2014, 111: 14147-14152. DOI:10.1073/pnas.1404171111 |
| [22] |
VYNNYTSKA-MYRONOVSKA B, KURLISHCHUK Y, BOBAK Y, DITTFELD C, KUNZ-SCHUGHART L A, STASYK O. Three-dimensional environment renders cancer cells profoundly less susceptible to a single amino acid starvation[J]. Amino Acids, 2013, 45: 1221-1230. DOI:10.1007/s00726-013-1586-x |
| [23] |
VYNNYTSKA-MYRONOVSKA B, BOBAK Y, GARBE Y, DITTFELD C, STASYK O, KUNZ-SCHUGHART L A. Single amino acid arginine starvation efficiently sensitizes cancer cells to canavanine treatment and irradiation[J]. Int J Cancer, 2012, 130: 2164-2175. DOI:10.1002/ijc.26221 |
| [24] |
VAN GELDERMALSEN M, QUEK L E, TURNER N, FREIDMAN N, PANG A, GUAN Y F, et al. Benzylserine inhibits breast cancer cell growth by disrupting intracellular amino acid homeostasis and triggering amino acid response pathways[J/OL]. BMC Cancer, 2018, 18: 689. DOI: 10.1186/s12885-018-4599-8.
|
| [25] |
VISSERS Y L, DEJONG C H, LUIKING Y C, FEARON K C, VON MEYENFELDT M F, DEUTZ N E. Plasma arginine concentrations are reduced in cancer patients: evidence for arginine deficiency?[J]. Am J Clin Nutr, 2005, 81: 1142-1146. DOI:10.1093/ajcn/81.5.1142 |
| [26] |
DELAGE B, FENNELL D A, NICHOLSON L, MCNEISH I, LEMOINE N R, CROOK T, et al. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer[J]. Int J Cancer, 2010, 126: 2762-2772. |
| [27] |
ENSOR C M, HOLTSBERG F W, BOMALASKI J S, CLARK M A. Pegylated arginine deiminase (ADI-SS PEG20, 000 mW) inhibits human melanomas and hepatocellular carcinomas in vitro and in vivo[J]. Cancer Res, 2002, 62: 5443-5450. |
| [28] |
LONG Y, TSAI W B, WANG D, HAWKE D H, SAVARAJ N, FEUN L G, et al. Argininosuccinate synthetase 1(ASS1) is a common metabolic marker of chemosensitivity for targeted arginine-and glutamine-starvation therapy[J]. Cancer Lett, 2017, 388: 54-63. DOI:10.1016/j.canlet.2016.11.028 |
| [29] |
MORRIS S M. Recent advances in arginine metabolism: roles and regulation of the arginases[J]. Br J Pharmacol, 2009, 157: 922-930. DOI:10.1111/j.1476-5381.2009.00278.x |
| [30] |
HU G D, WANG X, HAN Y, WANG P. Protein arginine methyltransferase 5 promotes bladder cancer growth through inhibiting NF-kB dependent apoptosis[J]. Excli J, 2018, 17: 1157-1166. |
| [31] |
ZHANG M, WU W, GAO M, ZHANG J, DING X, ZHU R, et al. Coactivator-associated arginine methyltransferase 1 promotes cell growth and is targeted by microRNA-195-5p in human colorectal cancer[J/OL]. Tumour Biol, 2017, 39: 1010428317694305. DOI: 10.1177/1010428317694305.
|
 2021, Vol. 42
2021, Vol. 42


