乳腺癌是女性最常见的恶性肿瘤之一,全球肿瘤估计数据显示,2018年约有63万人死于乳腺癌[1]。虽然近年来乳腺癌患者的生存率有所提高,但一旦出现复发或转移,生存时间将显著缩短[2]。因此,寻找乳腺癌发展过程中的新型标志物对改善乳腺癌的预后至关重要。此外,乳腺癌可根据雌激素受体(estrogen receptor,ER)、孕激素受体(progestogen receptor,PR)和人表皮生长因子受体2(human epidermal growth factor receptor 2,HER-2)的表达水平和扩增状态分为不同亚型,表达ER、PR和HER-2的患者预后好,治疗选择也更广泛,包括干预激素产生的内分泌治疗药物及抑制HER-2的靶向药物等[3-5];而ER、PR和HER-2均为阴性的三阴性乳腺癌(triple negative breast cancer,TNBC)恶性程度高,常发生于年轻女性,并且缺乏有效的靶向治疗药物[6-8]。TNBC对一些最有效的乳腺癌疗法不敏感,迫切需要寻找新的治疗靶点。
泛素化修饰属于蛋白质的翻译后修饰,在真核细胞的生物学过程中广泛存在。泛素系统已成为抗肿瘤药物研发的重要靶点,去泛素化酶抑制剂目前已进入临床试验阶段[9-10]。然而,去泛素化酶在肿瘤发生、发展过程中的信号调节过程仍不十分明确。泛素折叠修饰因子1特异性肽酶结构域蛋白(ubiquitin fold modifier 1-specific peptidase domain protein,ZUP1)是新近鉴定出来的第7种去泛素化酶,在复制叉停滞时期发挥重要作用。ZUP1不但能与复制体的重要组成部分相互识别,还可与复制叉上的DNA修复因子相互作用[11]。已有研究表明,人类肿瘤细胞中ZUP1的缺失将导致细胞内源性DNA损伤增加,这种内源性DNA损伤起源于细胞的S期[12]。总的来说,ZUP1在细胞应激反应相关的DNA复制过程中发挥着不可或缺的作用。然而,由于ZUP1是一种新型泛素化相关蛋白,其在肿瘤中的表达水平以及发挥的具体机制亟待探索。
本研究通过癌症基因组图谱(Cancer Genome Atlas,TCGA)等数据库分析ZUP1在乳腺癌中的表达水平及其与乳腺癌的临床病理参数及预后的关系,应用miRNA和泛素酶靶基因预测工具对潜在的ZUP1相关上游miRNA和泛素酶进行预测,通过基因集富集分析(gene set enrichment analysis,GSEA)研究ZUP1参与的下游信号通路,探讨ZUP1在乳腺癌发生、发展中的潜在分子机制。
1 资料和方法 1.1 微阵列数据集从基因表达汇编(Gene Expression Omnibus,GEO;http://www.ncbi.nlm.nih.gov/geo/)数据库下载乳腺癌相关阵列数据(GSE33692_GPL5175,GSE70947_GPL13607)。通过TCGA(https://portal.gdc.cancer.gov/)和基因型-组织表达(Genotype-Tissue Expression,GTEx;https://gtexportal.org/)数据库收集乳腺癌患者和正常对照组的临床资料和基因表达谱,检索并下载了1 104例乳腺癌患者和403例非癌症患者的数据集,以及TCGA中乳腺癌患者的临床病理数据(包括性别、年龄、种族、PAM50分型、病理分期、TNM分期、总生存期等)。
1.2 调控ZUP1的miRNA和泛素连接酶预测及下游基因富集分析采用在线工具TargetScan(http://www.targetscan.org/vert_72/)[13]、miRDB(http://www.mirdb.org/)[14]、miRTarBase(http://mirtarbase.mbc.nctu.edu.tw/php/index.php)[15]筛选以ZUP1为靶点的差异表达最大的前10个miRNA。使用UbiProber数据库(http://bioinfo.ncu.edu.cn/UbiProber.aspx)进行蛋白质泛素化预测[16]。采用GSEA程序(http://www.broadinstitute.org/gsea/index.jsp)分析TCGA乳腺癌队列的RNA序列数据[17],将标准化富集评分(normalized enrichment score,NES)绝对值>1且标称P<0.05作为筛选阈值。
1.3 统计学处理使用R语言edgeR包分析ZUP1基因在乳腺癌患者和正常对照组的表达差异。根据ZUP1基因表达水平中位数将样本分为高表达组和低表达组,采用χ2检验分析ZUP1基因表达水平与临床病理参数的关系。采用Kaplan-Meier法计算生存率,用log-rank检验比较ZUP1高表达组和低表达组生存率的差异[18]。
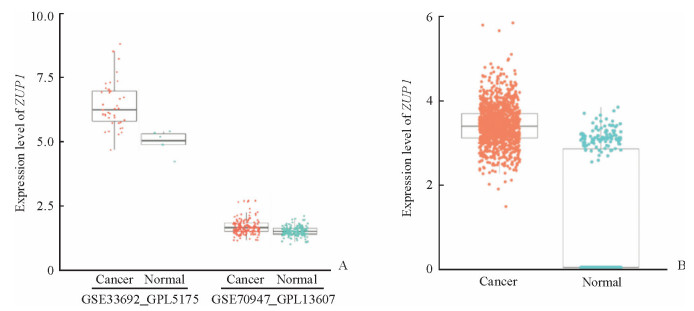
2 结果 2.1 ZUP1在乳腺癌组织中表达上调由图 1A可见,在GSE33692_GPL5175数据集中,乳腺癌患者的ZUP1基因表达高于正常对照组[log2FC=1.412,P<0.01;其中FC为差异表达倍数(fold change)];同样,在GSE70947_GPL13607数据集中,乳腺癌患者的ZUP1基因表达水平也高于正常对照组(log2FC=0.168,P<0.01)。由图 1B可见,对TCGA数据库及GTEx数据库中的1 104例乳腺癌组织和403例正常组织进行比较,ZUP1基因在乳腺癌患者癌组织中的表达也高于正常组织(log2FC=2.228,P<0.01)。
|
图 1 乳腺癌数据集中ZUP1基因的表达水平 Fig 1 Expression level of ZUP1 in breast cancer dataset A: The expression level of ZUP1 in breast cancer patients in the GSE33692_GPL5175 (48 tumor tissues and 8 normal tissues; log2FC=1.412, P < 0.01) and GSE70947_GPL13607 (148 pairs of tumor tissues and normal tissues; log2FC=0.168, P < 0.01) dataset; B: Expression level of ZUP1 in breast cancer and normal tissues in TCGA and GTEx databases (1 104 tumor tissues and 403 normal tissues; log2FC=2.228, P < 0.01). ZUP1: Ubiquitin fold modifier 1-specific peptidase domain protein; TCGA: Cancer Genome Atlas; GTEx: Genotype-Tissue Expression; FC: Fold change |
2.2 ZUP1基因表达水平与乳腺癌临床病理因素和预后的关系
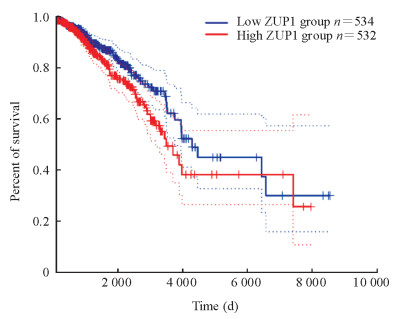
收集了1 090例乳腺癌患者ZUP1的临床病理资料,根据ZUP表达水平中位值分为高表达组和低表达组各545例。Kaplan-Meier生存分析结果(图 2)显示,ZUP1高表达患者比ZUP1低表达患者预后更差(HR=1.4,P=0.031)。由表 1可见,ZUP1基因的表达水平与乳腺癌病理T分期、PAM50分型、ER状态、PR状态、HER-2状态和组织学类型有关(P均<0.01)。
|
|
表 1 ZUP1的表达水平与乳腺癌患者临床病理因素的关系 Tab 1 Relationship between the expression of ZUP1 and clinicopathological factors of breast cancer patients |
|
图 2 ZUP1高表达组与低表达组乳腺癌患者的Kaplan-Meier生存分析 Fig 2 Kaplan-Meier survival analysis of breast cancer patients with high and low expression of ZUP1 The dotted line represents the 95% confidence interval. ZUP1: Ubiquitin fold modifier 1-specific peptidase domain protein |
2.3 ZUP1上游miRNA和泛素连接酶预测结果
生物信息学预测结果显示,以ZUP1基因为靶点的差异表达最大的前10个miRNA为miRNA-10b-3p、miRNA-499a-5p、miRNA-181b-2-3p、miRNA-181b-3p、miRNA-4420、miRNA-548aw、miRNA-5680、miRNA-570-3p、miRNA-7156-5p和miRNA-8087;可能调节ZUP1蛋白表达的泛素连接酶如表 2所示,其中评分居前5位的E3泛素连接酶包括膜相关RING-CH型蛋白(membrane-associated RING-CH,MARCH1)、MARCH8、鼠双微染色体2(mouse double minute 2,Mdm2)、滑膜蛋白(synoviolin)和E3泛素蛋白连接酶MIB1(mindbomb E3 ubiquitin protein ligase 1,MIB1)。
|
|
表 2 ZUP1蛋白相关泛素连接酶 Tab 2 ZUP1 protein-related ubiquitin ligase |
2.4 ZUP1下游基因集富集分析
共富集了50个功能基因集,明显富集上调前20个通路与下调的前14个通路分别见表 3、表 4,其中5条具有代表性的途径是基础转录因子、泛素介导的蛋白质水解、卵母细胞减数分裂、RNA降解和极光激酶B等通路。
|
|
表 3 GSEA分析结果中明显富集上调的前20个通路 Tab 3 The first 20 pathways significantly enriched and up-regulated in GSEA analysis |
|
|
表 4 GSEA分析结果中明显富集下调的前14个通路 Tab 4 The first 14 pathways significantly enriched and down-regulated in GSEA analysis |
3 讨论
泛素-蛋白酶体系统(ubiquitin-proteasome system,UPS)和溶酶体途径是真核细胞中蛋白降解的2条主要途径,分别负责降解短寿命蛋白和长寿命蛋白[19]。越来越多的证据表明,这2种降解途径之间并不完全独立,当USP活性受损时,溶酶体途径弥补了短寿命泛素化蛋白的降解[20]。既往研究表明,癌细胞会上调蛋白降解途径的相关蛋白,包括去泛素化酶等[21]。去泛素化酶是UPS降解的关键成分,负责在蛋白酶体降解之前去除泛素单体和泛素链。去泛素化酶家族成员已被证明在肿瘤微环境中存在差异表达和异常激活,且与肿瘤患者预后和治疗效果相关[22-23]。泛素连接酶和去泛素化酶也参与调控乳腺癌肿瘤细胞对化疗药物的敏感性,如E3连接酶F框唯一蛋白15(F-box only protein 15,FBXO15)和Casitas B谱系淋巴瘤原癌基因-b(Casitas B lymphoma-b,CBL-B)通过介导P-糖蛋白(P-glycoprotein,P-GP)的泛素化降解促进乳腺癌肿瘤细胞对药物的敏感性[24-25],去泛素化酶含卵巢肿瘤蛋白结构域的泛素乙酰结合蛋白1(ovarian tumor domain-containing ubiquitin aldehyde-binding protein 1,OTUB1)通过维持转录因子叉头框蛋白M1(forkhead box M1,FOXM1)的稳定促进乳腺癌化疗耐药[26]。为了发掘具有潜在作用的新型去泛素化酶,2018年2项研究同期报道了一类与其他已知的去泛素化酶家族没有同源性的新型半胱氨酸蛋白酶去泛素化酶,即ZUP1[27-28]。最初,ZUP1被认为是泛素折叠修饰因子1(ubiquitin fold modifier 1,UFM1)潜在的非活性蛋白酶[27]。尽管它的肽酶结构域与已知的UFM1蛋白酶有同源性,但是ZUP1缺乏切割UFM1所需的关键催化氨基酸残基。在结构上,ZUP1包含1个泛素结合域,在泛素介导的信号转导中具有重要作用[28]。为了证明ZUP1具有固有的去泛素化酶活性,Hewings等[27]发现ZUP1容易被泛素特异性活性探针所修饰,该探针能与去泛素化酶中的活性位点半胱氨酸残基发生不可逆反应;此外,他们还发现ZUP1对K63连接的多泛素链具有高度选择性。这一发现十分新颖,因为绝大多数去泛素化酶对于泛素链都是非选择性的,会切断大多数类型的多泛素连接结构。
目前仍鲜有研究提及ZUP1在肿瘤中的表达水平以及参与的相关分子机制。本研究通过TCGA、GEO和GTEx数据库检索收集乳腺癌患者的基因信息和临床病理数据,证明ZUP1在乳腺癌组织中表达上调,其表达水平与乳腺癌T分期、PAM50分型、ER状态、PR状态、HER-2状态和组织学类型有关,且高ZUP1水平的患者比低ZUP1水平的患者预后更差。生物信息学分析进一步表明,ZUP1与泛素介导的蛋白质水解、卵母细胞减数分裂、RNA降解和极光激酶B等通路有关。本研究预测了与ZUP1相关的多种E3泛素化蛋白连接酶,如MARCH1、MARCH8、Mdm2、synoviolin和MIB1;还预测了潜在调控ZUP1表达的miRNA,如miRNA-10b-3p、miRNA-499a-5p、miRNA-181b-2-3p、miRNA-181b-3p、miRNA-4420、miRNA-548aw、miRNA-5680、miRNA-570-3p、miRNA-7156-5p和miRNA-8087。研究发现,用姜黄素处理乳腺癌细胞后,miRNA-181b-2-3p水平变化最为显著;miRNA-181b-2-3p还可增强TNBC细胞MDA-MB-231细胞对多柔比星的敏感性,这是由于姜黄素通过激活活化T细胞核因子1(nuclear factor of activated T cells 1,NFAT1)的转录活性促进miRNA-181b-2-3p的表达[29]。因此,miRNA-181b-2-3p可能预测化疗反应,并可能成为肿瘤细胞对化疗药物致敏的治疗靶点。但是miRNA-181b-2-3p是否通过ZUP1发挥作用有待进一步探索。另有研究发现miRNA-570-3p在TNBC组织中的表达水平显著低于癌旁的正常组织,miRNA-570-3p能够靶向CD274抑制TNBC细胞的增殖、侵袭、迁移,诱导细胞凋亡,可能与抑制PI3K/AKT/mTOR信号通路有关[30]。尽管本研究预测了miRNA-181b-2-3p和miRNA-570-3p与ZUP1的表达有关,但其是否通过ZUP1发挥作用仍有待进一步探索。
ZUP1通过限制DNA损伤部位K63连接的多聚体链的形成,在调节基因组稳定通路中起着重要作用。在ZUP1的研究中仍然存在几个悬而未决的问题,例如ZUP1的底物是什么?为什么ZUP1更倾向于催化K63连接的泛素链?ZUP1缺失与特定DNA损伤反应途径有协同作用吗?ZUP1对K63连接的多聚体具有很高的选择性,因此它可能在复制应激后调节K63泛素化;或者,ZUP1可通过泛素结合锌指(ubiquitin-binding zinc finger,UBZ)结构域招募到在损伤部位形成的K6、K48或K63链,随后将K63链从基底上剥离。作为ZUP1活性的关键部位,UBZ结构域可能通过与多聚泛素结合介导底物募集,并作为S10位点使K63连锁链断裂。这些推测仍需进一步研究证实。
本研究发现ZUP1在乳腺癌中表达上调,并且与患者预后具有相关性,可能成为乳腺癌患者的一种新的预后生物标志物。ZUP1在乳腺癌发生、发展中的上下游机制可能与多种miRNA和多条信号通路有关,今后需要对大样本进行更详细的研究,以充分阐明ZUP1在乳腺癌中的作用。
| [1] |
BRAY F, FERLAY J, SOERJOMATARAM I, SIEGEL R L, TORRE L A, JEMAL A. Global cancer statistics 2018:GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2018, 68: 394-424. DOI:10.3322/caac.21492 |
| [2] |
LEMANSKI C, BOURGIER C, DRAGHICI R, THEZENAS S, MOREL A, ROUANET P, et al. Intraoperative partial irradiation for highly selected patients with breast cancer:results of the INTRAOBS prospective study[J]. Cancer Radiother, 2020, 24: 114-119. DOI:10.1016/j.canrad.2020.01.007 |
| [3] |
HAGAN C R, LANGE C A. Molecular determinants of context-dependent progesterone receptor action in breast cancer[J/OL]. BMC Med, 2014, 12: 32. doi: 10.1186/1741-7015-12-32.
|
| [4] |
DANIEL A R, GAVIGLIO A L, KNUTSON T P, OSTRANDER J H, D'ASSORO A B, RAVINDRANATHAN P, et al. Progesterone receptor-B enhances estrogen responsiveness of breast cancer cells via scaffolding PELP1- and estrogen receptor-containing transcription complexes[J]. Oncogene, 2015, 34: 506-515. DOI:10.1038/onc.2013.579 |
| [5] |
KNUTSON T P, LANGE C A. Tracking progesterone receptor-mediated actions in breast cancer[J]. Pharmacol Ther, 2014, 142: 114-125. DOI:10.1016/j.pharmthera.2013.11.010 |
| [6] |
ANDRÉ F, ZIELINSKI C C. Optimal strategies for the treatment of metastatic triple-negative breast cancer with currently approved agents[J/OL]. Ann Oncol, 2012, 23(Suppl 6): Ⅵ 46-Ⅵ 51. doi: 10.1093/annonc/mds195.
|
| [7] |
CHAVEZ K J, GARIMELLA S V, LIPKOWITZ S. Triple negative breast cancer cell lines:one tool in the search for better treatment of triple negative breast cancer[J]. Breast Dis, 2010, 32(1/2): 35-48. |
| [8] |
ANDERS C K, CAREY L A. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer[J]. Clin Breast Cancer, 2009, 9(Suppl 2): S73-S81. |
| [9] |
PRESTA I, NOVELLINO F, DONATO A, LA TORRE D, PALLERIA C, RUSSO E, et al. UbcH10 a major actor in cancerogenesis and a potential tool for diagnosis and therapy[J/OL]. Int J Mol Sci, 2020, 21: 2041. doi: 10.3390/ijms21062041.
|
| [10] |
YAO H, XU J. Regulation of cancer immune checkpoint:mono- and poly-ubiquitination:tags for fate[J]. Adv Exp Med Biol, 2020, 1248: 295-324. |
| [11] |
HAAHR P, BORGERMANN N, GUO X, TYPAS D, ACHUTHANKUTTY D, HOFFMANN S, et al. ZUFSP deubiquitylates K63-linked polyubiquitin chains to promote genome stability[J/OL]. Mol Cell, 2018, 70: 165-174.e6. doi: 10.1016/j.molcel.2018.02.024.
|
| [12] |
KWASNA D, ABDUL REHMAN S A, NATARAJAN J, MATTHEWS S, MADDEN R, DE CESARE V, et al. Discovery and characterization of ZUFSP/ZUP1, a distinct deubiquitinase class important for genome stability[J/OL]. Mol Cell, 2018, 70: 150-164.e6. doi: 10.1016/j.molcel.2018.02.023.
|
| [13] |
MON-LÓPEZ D, TEJERO-GONZÁLEZ C M. Validity and reliability of the TargetScan ISSF Pistol & Rifle application for measuring shooting performance[J]. Scand J Med Sci Sports, 2019, 29: 1707-1712. DOI:10.1111/sms.13515 |
| [14] |
CHEN Y, WANG X. miRDB: an online database for prediction of functional microRNA targets[J/OL]. Nucleic Acids Res, 2020, 48(D1): D127-D131. doi: 10.1093/nar/gkz757.
|
| [15] |
CHOU C H, CHANG N W, SHRESTHA S, HSU S D, LIN Y L, LEE W H, et al. miRTarBase 2016:updates to the experimentally validated miRNA-target interactions database[J]. Nucleic Acids Res, 2016, 44(D1): D239-D247. DOI:10.1093/nar/gkv1258 |
| [16] |
CHEN X, QIU J D, SHI S P, SUO S B, HUANG S Y, LIANG R P. Incorporating key position and amino acid residue features to identify general and species-specific ubiquitin conjugation sites[J]. Bioinformatics, 2013, 29: 1614-1622. DOI:10.1093/bioinformatics/btt196 |
| [17] |
POWERS R K, GOODSPEED A, PIELKE-LOMBARDO H, TAN A C, COSTELLO J C. GSEAInContext: identifying novel and common patterns in expression experiments[J/OL]. Bioinformatics, 2018, 34: i555-i564. doi: 10.1093/bioinformatics/bty271.
|
| [18] |
SALUJA R, CHENG S, DELOS SANTOS K A, CHAN K K W. Estimating hazard ratios from published KaplanMeier survival curves:a methods validation study[J]. Res Synth Methods, 2019, 10: 465-475. DOI:10.1002/jrsm.1362 |
| [19] |
CIECHANOVER A. Intracellular protein degradation:from a vague idea thru the lysosome and the ubiquitinproteasome system and onto human diseases and drug targeting[J]. Biochim Biophys Acta, 2012, 1824: 3-13. DOI:10.1016/j.bbapap.2011.03.007 |
| [20] |
LIN Z, BAZZARO M, WANG M C, CHAN K C, PENG S, RODEN R B. Combination of proteasome and HDAC inhibitors for uterine cervical cancer treatment[J]. Clin Cancer Res, 2009, 15: 570-577. DOI:10.1158/1078-0432.CCR-08-1813 |
| [21] |
BAZZARO M, LEE M K, ZOSO A, STIRLING W L, SANTILLAN A, SHIH IeM, et al. Ubiquitin-proteasome system stress sensitizes ovarian cancer to proteasome inhibitor-induced apoptosis[J]. Cancer Res, 2006, 66: 3754-3763. DOI:10.1158/0008-5472.CAN-05-2321 |
| [22] |
TIAN Z, D'ARCY P, WANG X, RAY A, TAI Y T, HU Y, et al. A novel small molecule inhibitor of deubiquitylating enzyme USP14 and UCHL5 induces apoptosis in multiple myeloma and overcomes bortezomib resistance[J]. Blood, 2014, 123: 706-716. DOI:10.1182/blood-2013-05-500033 |
| [23] |
LUISE C, CAPRA M, DONZELLI M, MAZZAROL G, JODICE M G, NUCIFORO P, et al. An atlas of altered expression of deubiquitinating enzymes in human cancer[J/OL]. PLoS One, 2011, 6: e15891. doi: 10.1371/journal.pone.0015891.
|
| [24] |
ZHANG Y, QU X, HU X, YANG X, HOU K, TENG Y, et al. Reversal of P-glycoprotein-mediated multi-drug resistance by the E3 ubiquitin ligase Cbl-b in human gastric adenocarcinoma cells[J]. J Pathol, 2009, 218: 248-255. DOI:10.1002/path.2533 |
| [25] |
KATAYAMA K, NOGUCHI K, SUGIMOTO Y. FBXO15 regulates P-glycoprotein/ABCB1 expression through the ubiquitin-proteasome pathway in cancer cells[J]. Cancer Sci, 2013, 104: 694-702. DOI:10.1111/cas.12145 |
| [26] |
KARUNARATHNA U, KONGSEMA M, ZONA S, GONG C, CABRERA E, GOMES A R, et al. OTUB1 inhibits the ubiquitination and degradation of FOXM1 in breast cancer and epirubicin resistance[J]. Oncogene, 2016, 35: 1433-1444. DOI:10.1038/onc.2015.208 |
| [27] |
HEWINGS D S, HEIDEKER J, MA T P, AHYOUNG A P, EL OUALID F, AMORE A, et al. Reactive-sitecentric chemoproteomics identifies a distinct class of deubiquitinase enzymes[J/OL]. Nat Commun, 2018, 9: 1162. doi: 10.1038/s41467-018-03511-6.
|
| [28] |
HERMANNS T, PICHLO C, WOIWODE I, KLOPFFLEISCH K, WITTING K F, OVAA H, et al. A family of unconventional deubiquitinases with modular chain specificity determinants[J/OL]. Nat Commun, 2018, 9: 799. doi: 10.1038/s41467-018-03148-5.
|
| [29] |
ZENG C, FAN D, XU Y, LI X, YUAN J, YANG Q, et al. Curcumol enhances the sensitivity of doxorubicin in triple-negative breast cancer via regulating the miR-181b-2-3p-ABCC3 axis[J/OL]. Biochem Pharmacol, 2020, 174: 113795. doi: 10.1016/j.bcp.2020.113795.
|
| [30] |
WANG L L, HUANG W W, HUANG J, HUANG R F, LI N N, HONG Y, et al. Protective effect of hsa-miR-570-3p targeting CD274 on triple negative breast cancer by blocking PI3K/AKT/mTOR signaling pathway[J]. Kaohsiung J Med Sci, 2020, 36: 581-591. DOI:10.1002/kjm2.12212 |
 2020, Vol. 41
2020, Vol. 41


