2. 重庆市铜梁区人民医院肿瘤血液内科, 重庆 402560
2. Department of Oncology and Hematology, The People's Hospital of Tongliang District, Chongqing 402560, China
肝细胞癌(hepatocellular carcinoma,HCC)是最常见的原发性肝脏恶性肿瘤,是世界范围内严重的健康问题[1]。它是全球第五大常见癌症和第三大癌症相关死亡原因[2]。转移是HCC患者死亡的主要原因。环状RNA(circular RNA,circRNA)是一类新的内源性非编码RNA,能够调节哺乳动物的基因表达。与线性RNA不同,circRNA具有共价闭环结构,没有5′-3′极性,也没有多聚腺苷酸尾[3-4]。circRNA的表达稳定、丰富、保守,通常在特定组织或特定发育阶段表达[5-6]。最近的研究表明,circRNA可能在许多生理和病理生理过程中发挥重要作用[7]。更多的研究表明,circRNA可以作为微RNA(microRNA,miRNA)的海绵,通过与miRNA相互作用调节基因表达[8-9]。miRNA的失调和功能在几乎所有生物过程中均已得到了广泛研究,但新鉴定的circRNA的表达和功能还需进一步研究。研究证实,circRNA失调(如circHIPK3和circSETD3)可能在HCC进展中发挥重要作用,直接调控HCC的细胞增殖、凋亡和迁移[10-11]。本研究拟分析circRNA circSP3(hsa-circ-0002642)在HCC患者癌组织和癌旁组织中的表达特点及其在正常肝细胞系和HCC细胞系中的表达特点,揭示circSP3对HCC细胞增殖、上皮-间质转化(epithelial-mesenchymal transformation,EMT)、迁移和侵袭的影响。
1 材料和方法 1.1 患者资料42例HCC患者的癌组织及配对癌旁组织(距离癌组织边缘>3 cm)均为重庆医科大学附属第一医院2017年6月至2018年12月收集的HCC手术切除标本。所有组织在切除后立即放入RNA保存液中,然后放置于- 80 ℃冰箱备用。所有患者术前均未进行化学治疗和放射治疗,且术后病理证实为HCC。其中男34例、女8例,国际抗癌联盟TNM分期Ⅰ ~ Ⅱ期20例、Ⅲ ~ Ⅳ期22例。所有患者及其家属均签署知情同意书。本研究通过重庆医科大学附属第一医院伦理委员会审批,并符合2013年《赫尔辛基宣言》的伦理准则。
1.2 细胞株及主要试剂人HCC细胞系Hep-3B、Huh7、SMMC-7721、Bel-7402及正常肝细胞系HL-7702均由重庆医科大学病毒性肝炎研究所保存。DMEM培养液和RPMI 1640培养液均购自美国Gibco公司;胎牛血清购自美国PAN公司;TRIzol和Lipofectamine 2000均购自美国Invitrogen公司;circRNA反转录及实时定量PCR试剂盒均购自广州吉赛生物科技股份有限公司;circSP3 PCR引物购自北京擎科生物技术有限公司;细胞计数试剂盒-8(cell counting kit-8,CCK-8)购自汉恒生物科技(上海)有限公司;circSP3过表达(ov-circSP3)及干扰质粒(si-circSP3)均购自吉玛基因(上海)股份有限公司。对于circSP3过表达,将circSP3的全长编码序列克隆到pcDNA3.1(+)载体中,空白pcDNA3.1(+)载体用作阴性对照[12]。si-circSP3序列为5′-GUC CUG CAG ACA GGU GAU UTT-3′,阴性对照si-NC序列为5′-UUC UCC GAA CGU GUC ACG UTT-3′。Transwell小室购自美国Corning公司;总蛋白提取试剂盒、BCA蛋白定量试剂盒、十二烷基硫酸钠-聚丙烯酰胺凝胶配制试剂盒和聚偏二氟乙烯膜均购自上海碧云天生物技术有限公司;兔抗波形蛋白(vimentin)、兔抗E-钙黏蛋白(E-cadherin)和兔抗甘油醛-3-磷酸脱氢酶(glyceraldehyde-3-phosphate dehydrogenase,GAPDH)单克隆抗体及羊抗兔二抗均购自北京博奥森生物技术有限公司;ECL试剂盒购自美国Advansta公司。
1.3 细胞培养及转染Hep-3B、Huh-7细胞培养于含10%胎牛血清的DMEM培养液中,SMMC-7721、Bel-7402细胞及正常肝细胞系HL-7702培养于含10%胎牛血清的RPMI 1640培养液中,细胞放置于37 ℃、5% CO2培养箱内培养,当细胞生长至贴壁达80% ~ 90%时进行传代。在质粒转染实验中,将处于对数生长期的细胞使用胰酶消化并计数、铺板。选用Huh-7细胞进行circSP3干扰及对照空载质粒转染,另外选用Hep-3B细胞进行circSP3过表达及对照空载质粒转染。转染前1 d将细胞接种于6孔板中,当细胞融合度达到50%左右时,按照转染试剂Lipofectamine 2000说明书的步骤进行转染。
1.4 实时定量PCR检测circSP3表达使用TRIzol试剂提取组织或细胞总RNA。反转录合成cDNA。以cDNA为模板,使用circRNA实时定量PCR试剂盒进行PCR。引物序列:circSP3正义引物5′-CTC CAG TTA GTC TAA GCA CTG G-3′,反义引物5′-GCC AAA TCA CCT GTC TGC AG-3′;内参照GAPDH正义引物5′-CAG CTA GCC GCA TCT TCT TTT-3′,反义引物5′-GTG ACC AGG CGC CCA ATA C-3′。每个样品设3个复孔,采用2- ΔΔCt法计算circSP3的相对表达量。
1.5 Transwell法检测细胞侵袭、迁移能力细胞侵袭能力检测实验:溶解基底胶(按照1 : 9稀释基底胶),向小室上层每孔加入50 μL基底胶。细胞转染48 h后进行消化、计数,取约5×104个细胞重悬于200 μL无血清DMEM培养液,加入Transwell上室。向下室加入500 μL含10%胎牛血清的DMEM培养液,然后置于37 ℃、5% CO2培养箱中孵育24 h。取出小室,用磷酸盐缓冲液清洗3次,加入4%多聚甲醛溶液固定30 min,用棉棒擦拭清除上室未通过的细胞,然后用结晶紫染色20 min,磷酸盐缓冲液清洗3次。使用光学显微镜于20倍对下室细胞进行计数及拍照。细胞迁移能力检测实验:Transwell上室不铺基底胶,其余步骤与侵袭能力检测实验相同。
1.6 蛋白质印迹法检测EMT相关蛋白表达使用裂解液充分裂解细胞并提取细胞总蛋白,加入蛋白上样缓冲液混合,沸水煮10 min。每孔上样15 μg蛋白质样品,电泳(80 V 40 min、120 V 80 min)、转膜(210 mA 100 min)。室温下用8%脱脂牛奶封闭2 h,然后4 ℃过夜标记波形蛋白、E-钙黏蛋白(稀释比例均为1 : 1 000)和GAPDH(1 : 800)一抗;然后加入羊抗兔二抗(稀释比例1 : 1 000)室温孵育2 h。使用ECL试剂盒进行化学发光显影,用ImageJ软件进行半定量分析。
1.7 统计学处理应用SPSS 24.0软件进行统计学分析。所有实验均独立重复3次,计量资料以x±s表示,2组间比较采用t检验,多组间比较采用单因素方差分析。circSP3表达与患者临床病理指标的关系采用χ2检验进行分析。检验水准(α)为0.05。
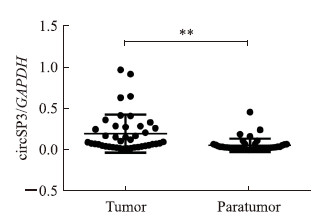
2 结果 2.1 circSP3在癌及癌旁组织中的表达及其与临床病理指标的关系如图 1所示,癌组织中circSP3的表达水平高于癌旁组织(P<0.01)。以癌组织circSP3相对表达量的平均值为界,23例患者circSP3高表达,19例患者为低表达。由表 1可见,肿瘤最大径≥ 5 cm和高TNM分期的HCC患者多表现为circSP3高表达(P均<0.05)。表明circSP3可能参与HCC的形成,且circSP3表达水平与HCC病情的严重程度呈正相关。
|
图 1 实时定量PCR检测HCC癌组织和癌旁组织中circSP3表达 Fig 1 Expression of circSP3 in HCC tumor and paratumor tissues detected by real-time quantitative PCR HCC: Hepatocellular carcinoma; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase. **P < 0.01. n = 42 |
|
|
表 1 circSP3表达与HCC患者临床病理指标的相关性 Tab 1 Correlation between circSP3 expression and clinicopathological characteristics of HCC patients |
2.2 circSP3在人HCC细胞系及正常肝细胞系中的表达
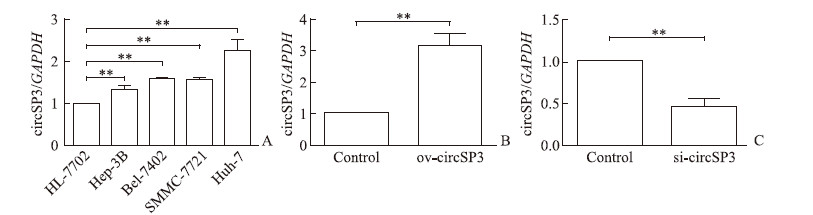
如图 2A所示,与HL-7702细胞相比,Huh-7、Hep-3B、SMMC-7721、Bel-7402细胞中circSP3表达均上调(P均<0.01)。在Hep-3B和Huh-7细胞中分别转染circSP3过表达和干扰质粒后,circSP3的表达被成功上调和下调(P均<0.01,图 2B、2C)。
|
图 2 实时定量PCR检测各HCC细胞中circSP3表达及转染ov-circSP3和si-circSP3质粒后circSP3表达 Fig 2 Expression of circSP3 in HCC cells after transfection with ov-circSP3 and si-circSP3 plasmids detected by real-time quantitative PCR A: The expression of circSP3 in human HCC cell lines (Huh-7, Hep-3B, SMMC-7721 and Bel-7402) and human normal liver cell line (HL-7702); B: The expression of circSP3 in Hep-3B cells transfected with circSP3 overexpression plasmids (ov-circSP3) or empty plasmids (control); C: The expression of circSP3 in Huh-7 cells transfected with circSP3 interference plasmids (si-circSP3) or empty plasmids (control). HCC: Hepatocellular carcinoma; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase. **P < 0.01. n = 3, x±s |
2.3 circSP3对HCC细胞增殖的影响
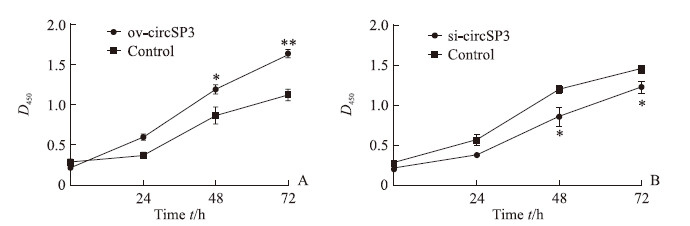
如图 3所示,转染circSP3过表达质粒的Hep-3B细胞增殖活性较对照细胞升高,在转染48、72 h时差异均有统计学意义(P均<0.05),而转染了circSP3干扰质粒的Huh-7细胞的增殖活性较对照细胞降低,在转染48、72 h时差异亦均有统计学意义(P均<0.05)。
|
图 3 CCK-8法检测circSP3对HCC细胞增殖能力的影响 Fig 3 Effect of circSP3 on proliferation of HCC cells detected by CCK-8 method A: The proliferation of Hep-3B cells transfected with circSP3 overexpression plasmids (ov-circSP3) or empty plasmids (control); B: The proliferation of Huh-7 cells transfected with circSP3 interference plasmids (si-circSP3) or empty plasmids (control). CCK-8: Cell counting kit-8; HCC: Hepatocellular carcinoma. *P < 0.05, **P < 0.01 vs control group at same time point. n = 3, x±s |
2.4 circSP3对HCC细胞侵袭、迁移的影响
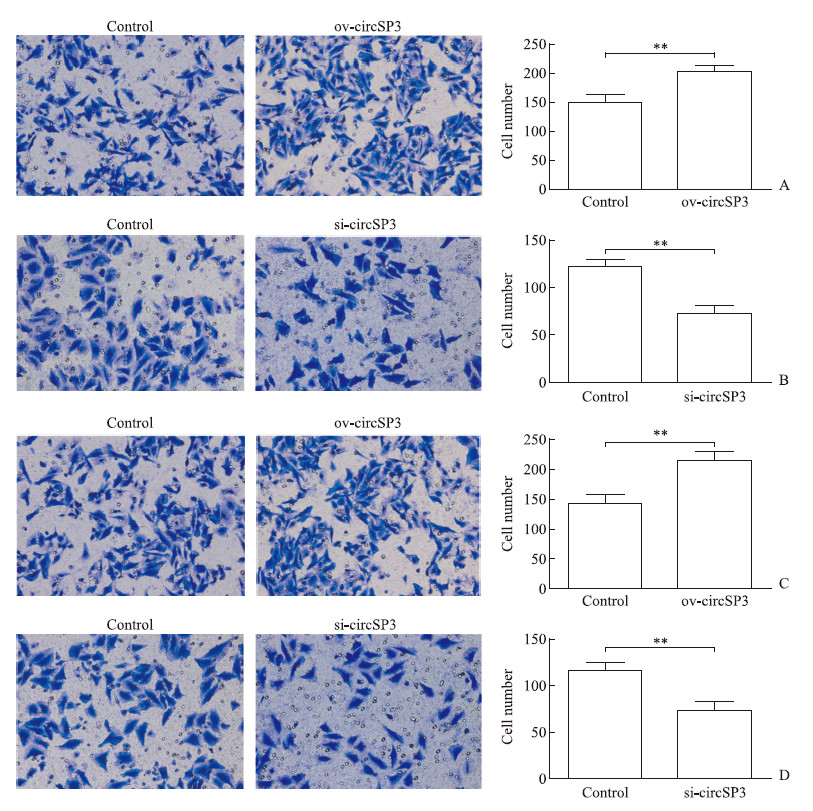
如图 4A、4B所示,与对照组相比,circSP3过表达组穿过小室的细胞数增多(P<0.01),circSP3干扰组穿过小室的细胞数减少(P<0.01),表明circSP3表达能够促进HCC细胞的侵袭能力。如图 4C、4D所示,circSP3表达亦能够促进HCC细胞的迁移能力(P<0.01)。
|
图 4 Transwell法检测circSP3对HCC细胞侵袭、迁移的影响 Fig 4 Effect of circSP3 on invasion and migration of HCC cells detected by Transwell assay A: The invasion of Hep-3B cells transfected with circSP3 overexpression plasmids (ov-circSP3) or empty plasmids (control); B: The invasion of Huh-7 cells transfected with circSP3 interference plasmids (si-circSP3) or empty plasmids (control); C: The migration of Hep-3B cells transfected with ov-circSP3 or control plasmids; D: The migration of Huh-7 cells transfected with si-circSP3 or control plasmids. HCC: Hepatocellular carcinoma. Original magnification: ×20. **P < 0.01. n = 3, x±s |
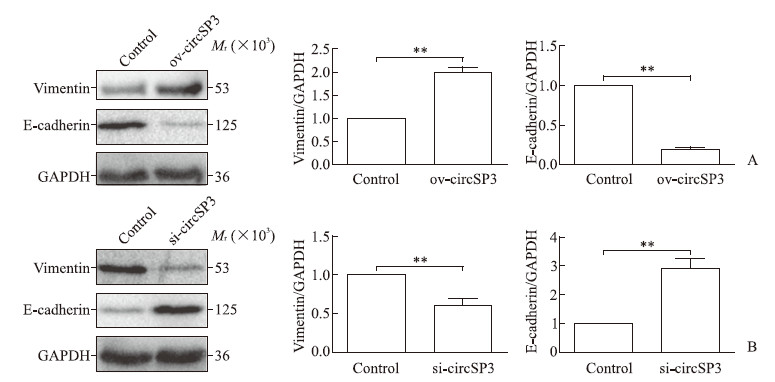
2.5 circSP3调节EMT相关蛋白波形蛋白、E-钙黏蛋白表达
如图 5所示,与对照组相比,circSP3过表达组Hep-3B细胞中波形蛋白的表达水平升高(P<0.01),E-钙黏蛋白表达水平降低(P<0.01);circSP3干扰组Huh-7细胞中波形蛋白的表达水平降低(P<0.01),E-钙黏蛋白表达水平升高(P<0.01)。这些结果表明,circSP3表达能促进EMT过程。
|
图 5 蛋白质印迹法检测circSP3对HCC细胞中EMT相关蛋白波形蛋白和E-钙黏蛋白表达的影响 Fig 5 Effect of circSP3 on expression of EMT-related proteins vimentin and E-cadherin in HCC cells detected by Western blotting A: The protein expression in Hep-3B cells transfected with circSP3 overexpression plasmids (ov-circSP3) or empty plasmids (control); B: The protein expression in Huh-7 cells transfected with circSP3 interference plasmids (si-circSP3) or empty plasmids (control). HCC: Hepatocellular carcinoma; EMT: Epithelial-mesenchymal transformation; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase. **P < 0.01. n = 3, x±s |
3 讨论
circRNA是一类新的非编码RNA成员,包括外显子circRNA、内含子circRNA和外显子-内含子circRNA [13-14]。与已知的线性RNA不同,circRNA是封闭的共价结合连续环结构,既不具有5′-3′极性又不具有多聚腺苷酸尾,这阻止了它们被RNA外切核酸酶或RNA酶R降解[15]。因此,circRNA比线性mRNA更稳定,这使得它们比来自相同基因的典型线性转录本更丰富[6]。此外,circRNA能够在转录或转录后水平调节基因表达。
外显子circRNA组成大部分的circRNA,主要存在于细胞质,并可能作为miRNA海绵而发挥作用;而内含子circRNA和外显子-内含子circRNA在细胞核中含量更丰富,并可能调控基因转录和转录后水平[13, 16-17]。此外,已有研究发现它们存在于不同的细胞外液中,包括唾液、血液和尿液[18]。这些研究结果提示circRNA可能参与不同的生物学过程,如细胞增殖、侵袭和迁移、凋亡和自噬等。
EMT是上皮细胞失去极性、经过细胞骨架重塑转变成具有迁移能力的间充质表型的过程。多项研究证实EMT是肿瘤侵袭转移的一个重要步骤[19],且EMT被认为是肿瘤转移的标志[20],可以增加HCC细胞的迁移及侵袭能力[21],其主要的特征为通过Wnt和Notch信号通路使E-钙黏蛋白表达减少、波形蛋白表达增加。
通过StarBase数据库分析可以确定circSP3(chr2:174819600–174820960)来自于SP3基因座中的外显子区域,由5个外显子成环组成,该区域位于2q31.1号染色体上。本研究结果显示,与正常肝细胞系相比,circSP3表达在4种HCC细胞中均上调。同时,在42例HCC患者中,癌组织中circSP3表达高于癌旁组织,并且具有更高TNM分期或更大肿瘤(≥ 5 cm)的患者circSP3表达水平更高,说明circSP3与HCC密切相关,并有作为诊断HCC生物标志物的潜力。本研究结果显示,circSP3过表达能够促进HCC细胞增殖,干扰circSP3表达则可抑制HCC细胞的增殖;并且circSP3过表达能促进HCC细胞的侵袭及迁移,干扰circSP3表达能抑制HCC细胞的侵袭及迁移。因此circSP3可能是促进HCC转移的潜在分子标志物。同时,过表达及干扰circSP3表达后检测EMT相关蛋白E-钙黏蛋白、波形蛋白的表达量变化,证明circSP3表达能促进EMT进程。上述结果提示circSP3可能通过促进EMT进程而促进HCC细胞的迁移和侵袭,在HCC进展中发挥重要作用。
| [1] |
KHATTAB M, FOUAD M, AHMED E. Role of biomarkers in the prediction and diagnosis of hepatocellular carcinoma[J]. World J Hepatol, 2015, 7: 2474-2481. DOI:10.4254/wjh.v7.i23.2474 |
| [2] |
CALDWELL S, PARK S H. The epidemiology of hepatocellular cancer:from the perspectives of public health problem to tumor biology[J]. J Gastroenterol, 2009, 44(Suppl 19): 96-101. |
| [3] |
LLOVET J M, ZUCMAN-ROSSI J, PIKARSKY E, SANGRO B, SCHWARTZ M, SHERMAN M, et al. Hepatocellular carcinoma[J/OL]. Nat Rev Dis Primers, 2016, 2: 16018. doi: 10.1038/nrdp.2016.18.
|
| [4] |
TORRE L A, BRAY F, SIEGEL R L, FERLAY J, LORTET-TIEULENT J, JEMAL A. Global cancer statistics, 2012[J]. CA Cancer J Clin, 2015, 65: 87-108. DOI:10.3322/caac.21262 |
| [5] |
HANSEN T B, JENSEN T I, CLAUSEN B H, BRAMSEN J B, FINSEN B, DAMGAARD C K, et al. Natural RNA circles function as efficient microRNA sponges[J]. Nature, 2013, 495: 384-388. DOI:10.1038/nature11993 |
| [6] |
JECK W R, SORRENTINO J A, WANG K, SLEVIN M K, BURD C E, LIU J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats[J]. RNA, 2013, 19: 141-157. DOI:10.1261/rna.035667.112 |
| [7] |
QIU L P, WU Y H, YU X F, TANG Q, CHEN L, CHEN K P. The emerging role of circular RNAs in hepatocellular carcinoma[J]. J Cancer, 2018, 9: 1548-1559. DOI:10.7150/jca.24566 |
| [8] |
HAN D, LI J, WANG H, SU X, HOU J, GU Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression[J]. Hepatology, 2017, 66: 1151-1164. DOI:10.1002/hep.29270 |
| [9] |
HUANG X Y, HUANG Z L, XU Y H, ZHENG Q, CHEN Z, SONG W, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma[J/OL]. Sci Rep, 2017, 7: 5428. doi: 10.1038/s41598-017-05432-8.
|
| [10] |
CHEN G, SHI Y, LIU M, SUN J. CircHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma[J/OL]. Cell Death Dis, 2018, 9: 175. doi: 10.1038/s41419-017-0204-3.
|
| [11] |
XU L, FENG X, HAO X, WANG P, ZHANG Y, ZHENG X, et al. CircSETD3(Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth[J/OL]. J Exp Clin Cancer Res, 2019, 38: 98. doi: 10.1186/s13046-019-1041-2.
|
| [12] |
WESSELHOEFT R A, KOWALSKI P S, ANDERSON D G. Engineering circular RNA for potent and stable translation in eukaryotic cells[J/OL]. Nat Commun, 2018, 9: 2629. doi: 10.1038/s41467-018-05096-6.
|
| [13] |
SALZMAN J, CHEN R E, OLSEN M N, WANG P L, BORWN P O. Cell-type specific features of circular RNA expression[J/OL]. PLoS Genet, 2013, 9: e1003777. doi: 10.1371/journal.pgen.1003777.
|
| [14] |
ZHANG Y, ZHANG X O, CHEN T, XIANG J F, YIN Q F, XING Y H, et al. Circular intronic long noncoding RNAs[J]. Mol Cell, 2013, 51: 792-806. DOI:10.1016/j.molcel.2013.08.017 |
| [15] |
SUZUKI H, TSUKAHARA T. A view of pre-mRNA splicing from RNase R resistant RNAs[J]. Int J Mol Sci, 2014, 15: 9331-9342. DOI:10.3390/ijms15069331 |
| [16] |
MEMCZAK S, JENS M, ELEFSINIOTI A, TORTI F, KRUEGER J, RYBAK A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency[J]. Nature, 2013, 495: 333-338. DOI:10.1038/nature11928 |
| [17] |
YU B, SHAN G. Functions of long noncoding RNAs in the nucleus[J]. Nucleus, 2016, 7: 155-166. DOI:10.1080/19491034.2016.1179408 |
| [18] |
QU S, YANG X, LI X, WANG J, GAO Y, SHANG R, et al. Circular RNA:a new star of noncoding RNAs[J]. Cancer Lett, 2015, 365: 141-148. DOI:10.1016/j.canlet.2015.06.003 |
| [19] |
MICALIZZI D S, FARABAUGH S M, FORD H L. Epithelial-mesenchymal transition in cancer:parallels between normal development and tumor progression[J]. J Mammary Gland Biol Neoplasia, 2010, 15: 117-134. DOI:10.1007/s10911-010-9178-9 |
| [20] |
KSIĄŻKIEWICZ M, MARKIEWICZ A, ZACZEK A J. Epithelial-mesenchymal transition:a hallmark in metastasis formation linking circulating tumor cells and cancer stem cells[J]. Pathobiology, 2012, 79: 195-208. DOI:10.1159/000337106 |
| [21] |
REICHL P, HAIDER C, GRUBINGER M, MIKULITS W. TGF-β in epithelial to mesenchymal transition and metastasis of liver carcinoma[J]. Curr Pharm Des, 2012, 18: 4135-4147. DOI:10.2174/138161212802430477 |
 2019, Vol. 40
2019, Vol. 40


