长链非编码RNA(long non-coding RNA,lncRNA)是指长度超过200 nt且通常不具备蛋白质编码功能的RNA(近年来的研究发现,部分lncRNA可以编码一些短肽,它们的具体功能尚不清楚[1])。得益于高通量测序技术的进步,近年来大量的lncRNA被发现,目前已知它们在生物界中普遍存在,根据最新的lncRNA数据库网站NONCODE第5版[2]资料,在包括真菌、植物、昆虫、线虫、脊椎动物在内的17种生物中已鉴定出超过54万个lncRNA转录本。在人类,无论就转录本的绝对数量还是种类多少而言,lncRNA都是一个庞大的类群。由于lncRNA是按长度定义的,因此它并不像tRNA、rRNA、mRNA那样有相对固定的功能模式,不同lncRNA在各细胞系中的表达以及在亚细胞层面的定位都呈现出特异性[3-5],提示其具有不同的生物学功能和作用机制,因此直接对特定lncRNA进行功能预测较为困难。为了充分了解这一群体在生理、病理过程中的作用,必须通过一系列独立实验探索各种lncRNA的功能并详细注释,而目前被有效注释的lncRNA只占一小部分[6]。在免疫学研究领域,对lncRNA如何参与天然免疫过程的理解正随着越来越多lncRNA功能的发现而不断深化。Guttman等[7]于2009年发现脂多糖刺激的小鼠骨髓来源树突状细胞诱导多种lncRNA的转录,率先揭示了lncRNA与天然免疫之间的联系。随着微阵列芯片和高通量RNA测序的广泛应用以及各种功能验证实验体系的建立与完善,已证明在不同细胞系中多种lncRNA对天然免疫具有重要的调控作用,包括决定天然免疫细胞分化、调控炎性细胞因子的表达以及参与病毒和宿主的相互作用等[8-14]。本文从lncRNA的特点、受调控的天然免疫效应、分子机制3个方面介绍lncRNA在天然免疫中的作用,并讨论天然免疫相关的lncRNA研究策略和技术进展。
1 LncRNA的特点与mRNA类似,大多数lncRNA也需要剪接,并且具有5′端帽和多聚腺苷酸尾结构[15]。但lncRNA与mRNA在生物学特性上差异明显,除了无蛋白编码能力,lncRNA还有以下4个方面特点。
(1)LncRNA的数量和种类繁多。过去大量的转录组分析表明,人类基因组中大约有2/3的基因会被转录,而其中大部分为非编码RNA,只有2%的RNA翻译蛋白[16]。据估计人类基因组中有20 000~25 000种蛋白编码基因[17],而NONCODE数据库收录的人类细胞lncRNA基因有90 000多个[2]。研究发现,越复杂和高级的生物体内lncRNA的种类越多,说明非编码RNA调控对生物有重要的进化学意义[18]。
(2)LncRNA具有众多生物学功能。LncRNA通过长度和编码能力定义,没有固定的功能模式,通过各种不同的机制发挥作用。已经发现的功能有:①转录前调控。调节信号通路,影响目标基因的表达[19]或影响转录因子与启动子结合,调节下游基因[11]。②转录后调控。参与对前体mRNA的剪接[20]、翻译[21]和降解[22]。③翻译后调控。特异性地与蛋白酶结合,改变其催化活性[23]。④表观遗传学修饰。参与基因印迹[24]、染色质重塑[25]等生物学过程。
(3)LncRNA可以折叠形成具有功能的高级结构。众所周知,mRNA编码区的一级结构是决定其编码能力的关键,突变可能导致翻译错误;而lncRNA一级结构保守性不强,因此曾有学者据此认为非编码RNA是转录过程中无生物学意义的“噪声”[26]。深入研究发现,lncRNA虽然总体保守性不强,但有一些保守性较强的元件,比如THCAT126含有存在于所有脊椎动物的保守元件[27],提示其功能和特定结构之间存在联系。目前认为lncRNA既能够通过碱基互补配对与核酸序列结合,也能通过二级或更高级的折叠结构与蛋白质相互作用[28]。
(4)LncRNA的分布在细胞和亚细胞层面均具有特异性。在细胞层面,许多lncRNA在特定的细胞系中高表达:有学者对人类15种细胞系进行转录组学分析发现,多达29%的lncRNA特异地表达于一个细胞系,只有10%的lncRNA在所有细胞系中表达,而mRNA上述2个比例分别为7%和53%[3];Liu等[4]的研究也得出同样结论,他们检测了7个人类细胞系中的1 329个lncRNA基因,筛选出499个生长关键基因,绝大多数(89%)只影响1种细胞的生长,并且所有检测的lncRNA没有一个存在于所有细胞系中。在亚细胞层面,对于相同种类的细胞,lncRNA同样呈现出特异的亚细胞定位,例如中性粒细胞中Morrbid定位于核内[29]、树突状细胞中lnc-DC定位于细胞质[10]。LncATLAS是专门提供lncRNA亚细胞定位检索的数据库,通过该数据库能够查阅到更多相关信息[5]。目前在筛选功能性lncRNA过程中必须考虑其亚细胞定位,因为定位往往为潜在功能提供了线索。
基于上述特点,不同的lncRNA和其他生物分子相互作用,共同构成一个复杂而精密的调控网络。
2 LncRNA在天然免疫中的效应目前认为,lncRNA广泛参与多种天然免疫效应,包括诱导天然免疫细胞的分化、调节炎性细胞因子的分泌、参与病毒等病原体和宿主的相互作用等[30-33]。
通过对不同分化阶段细胞转录组学差异倍数的比较,筛选出一些对天然免疫细胞的分化具有关键作用的lncRNA,例如促进单核细胞向巨噬细胞分化的lnc-MC[34],维持中性粒细胞、嗜酸性粒细胞存活的Morrbid[29],促使单核细胞分化为成熟树突状细胞的lnc-DC[10],分别维持1型和3型天然免疫淋巴细胞(innate lymphoid cell,ILC)谱系特征和功能的Rroid[35]和lncKdm2b[36]等。
在天然免疫调控方面,各种炎症相关lncRNA参与了对炎性细胞因子网络的精密调控。有的lncRNA起正向调控作用,例如PACER介导环氧化酶2(cyclooxygenase 2,COX-2)表达升高[37]、THRIL促进以肿瘤坏死因子(tumor neucrosis factor,TNF)为主的多种细胞因子及趋化因子的产生[8]、NEAT1使IL-8表达上升[38]、AS-IL-1α促进IL-1α的转录[39]等;还有一些lncRNA起负反馈抗炎作用,例如lincRNA-Cox2的缺失将导致500多种炎性因子表达上调[11]、Lethe负反馈抑制NF-κB的表达[9]、lncRNA-CMPK2下调数个干扰素刺激相关基因[40]、AS-IL-1β抑制IL-1β的表达[41]等。
功能性lncRNA参与了病毒[42]、细菌[43]、寄生虫[44]等病原体与宿主之间的作用。这种作用是相互的,以病毒为例,某些lncRNA帮助宿主发挥抗病毒的功能,例如NeST[43, 45]、NEAT1[46]、HULC[47-48]、EBER[49]等;而另一些lncRNA由病原体诱导宿主高表达或者直接由病原体编码,它们帮助病毒复制、削弱免疫系统或者逃避免疫防御[50],例如卡波西肉瘤相关疱疹病毒(Kaposi’s sarcoma-associated herpesvirus,KSHV)诱导的多聚腺苷酸化核(polyadenylated nuclear,PAN)RNA[51-52]、疱疹性口炎病毒感染时表达升高的lncRNA-ACOD1[23]等。另外,病毒也可以编码lncRNA,有几项独立的研究分别观察到一种由人类免疫缺陷病毒(human immunodeficiency virus,HIV)编码的反义lncRNA[53-55]。Saayman等[56]进一步研究发现这种lncRNA能够招募内源的染色体重塑蛋白Dnmt3a、HDAC1和EZH2.65使病毒染色体沉默、转录停止,而干扰该lncRNA能重新激活病毒的转录。病毒编码一种抑制自身的lncRNA似乎违背常理,但是研究者认为这可能与HIV的潜伏有关。无论如何,这个发现表明人们对于病毒和lncRNA相互作用的了解还不充分。表 1列举了已证实具有天然免疫效应的部分lncRNA的生物学功能及其分子作用机制。
|
|
表 1 天然免疫相关lncRNA生物学功能及其分子作用机制 Tab 1 Biological functions and molecular mechanisms of innate immunity-related lncRNA |
3 LncRNA在天然免疫系统中发挥功能的分子机制
为了更好地阐明lncRNA如何参与上述生物学效应,必须深入了解其分子机制。综合lncRNA的研究报道,目前lncRNA发挥功能的分子机制有以下8种(图 1)。
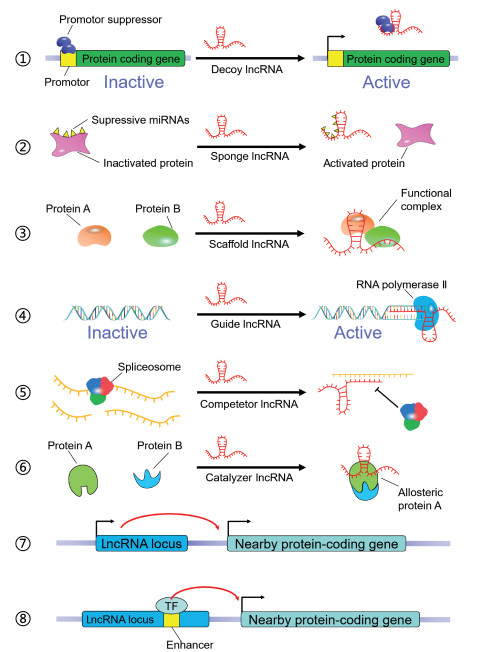
|
图 1 LncRNA发挥功能的分子机制 Fig 1 Molecular mechanisms of functional lncRNA ① Decoy: LncRNA combines biological molecules such as proteins in a titration manner, thus hinder their function; ② Sponge: LncRNA absorbs miRNAs, making them exhausted; ③ Scaffold: LncRNA connects two different proteins to make them a functional complex; ④ Guide: LncRNA recruits other molecules to a target to exert their functions; ⑤ Interacts with mRNA: LncRNA specifically connects with certain mRNA to interfere with its splicing activity; ⑥ Changes protein's structure: LncRNA directly connects with enzymes, changing their catalytic activities through allosteric effect; ⑦ Transcription of lncRNA locus: Transcription and splicing of lncRNA is necessary to induce expression of nearby protein-coding genes in some circumstances; ⑧ LncRNA locus may contains functional elements: The enhancers imbedded within lncRNA locus may be key to expression of nearby protein-coding genes. LncRNA: Long non-coding RNA; TF: Transcription factor |
3.1 分子诱饵
LncRNA与具有调节功能的蛋白通过类似“化学滴定中和”的方式结合后使其失去活性,产生抑制效果。例如PACER通过结合p50-p50二聚体,使后者对COX-2启动子区的抑制作用消失,启动COX-2基因的转录[37];Lethe与PACER的调控作用相反,通过结合p65-p65二聚体,阻断其在NF-κB、IL-6、IL-8等目标基因启动子区的聚集,从而限制这些炎症基因的表达[9];NEAT1不仅参与形成染色体旁斑[57],同时也是一种诱饵分子,当NEAT1表达升高时其结合脯氨酸/谷氨酰胺富含性剪切因子(splicing factor proline/glutamine-rich,SFPQ)形成旁斑,使得SFPQ对IL-8启动子的抑制作用消失,从而增强IL-8的表达[38]。
3.2 MicroRNA分子海绵与分子诱饵功能类似,分子海绵特指lncRNA结合大量的microRNA,使其衰竭而失去作用。例如lnc-MC通过大量吸收miR-199a-5p阻断miR-199a-5p对1B型激活素A受体(activin A receptor type 1B,ACVR1B)的抑制作用,由于ACVR1B是单核细胞分化的关键受体,因此lnc-MC间接激活了ACVR1B,使单核细胞向巨噬细胞分化[34]。
3.3 分子骨架LncRNA可以作为连接DNA、RNA、蛋白等生物分子的骨架,使它们组成有复杂活性的复合物。例如lncRNA RMRP可以促进T细胞RAR相关的孤儿核受体γ(RAR-related orphan receptor γ in thymocytes,RORγt)与死亡盒蛋白5(DEAD-box protein 5,DDX5)结合,RMRP-RORγt-DDX5复合体促使Th17细胞相关基因转录、原始CD4+ T细胞向Th17细胞分化并发挥功能,如表达IL-17a、IL-17f和IL-22等细胞因子[58]。
3.4 分子向导LncRNA还可以招募其他分子到作用靶点以发挥功能。例如Morrbid靠近Bcl2l11启动子区,结合并募集重要的组蛋白修饰复合物多梳蛋白抑制复合物2(polycomb repressive complex 2,PRC2),在Bcl2l11启动子上沉积H3K27me3组蛋白标记,下调Bcl2l11的表达[29];AS-IL-1α在IL-1α启动子区募集RNA聚合酶Ⅱ,引起IL-1α转录[39];lincRNA-p21能够招募核内不均一核糖核蛋白K(heterogeneous nuclear ribonucleoprotein K,hnRNP-K),结合到多种基因的启动子区,起到转录抑制作用,其中包括一些细胞存活关键基因,因此促使细胞凋亡[59]。
3.5 与mRNA相互作用LncRNA参与了对mRNA剪切、翻译、降解等生物学过程的调控。在剪切层面,一种被称为肺腺癌转移相关转录物1(metastasis-associated in lung adenocarcinoma transcript 1,MALAT1)的lncRNA在多种癌症中高表达,主要位于剪接斑点(splicing speckle)[60],它通过调控剪接因子在剪接斑点中的分布和磷酸化水平,改变前体mRNA的选择性剪接模式[61];在翻译层面,lincRNA-p21通过特异性结合Jun B、β连环蛋白(β-catenin)等蛋白的mRNA序列,形成RNA-RNA二聚体,阻止这些mRNA被翻译成蛋白质[21];在降解层面,一种被称为半Stau1(一种mRNA降解因子[62])结合位点RNA(half-Stau1-binding site RNA,1/2-sbsRNA)的lncRNA通过与mRNA 3′端非翻译区的Alu元件不完全配对形成Stau1结合位点,促进Stau1与mRNA结合,导致mRNA降解[63]。
3.6 蛋白变构效应LncRNA与多种酶类直接结合,包括代谢酶、激酶和各种表观修饰酶等,改变其催化活性,从而发挥多种调控功能。2017年12月本研究团队发现,在多种病毒诱导下一个特异的lncRNA lncRNA-ACOD1表达显著升高,其直接结合细胞质中的天冬氨酸转氨酶2(glutamic-oxaloacetic transaminase 2,GOT2),使得底物的作用位点的构象改变,进一步提高酶催化活性,显著影响细胞的代谢通路,从而促进病毒的复制[23]。
3.7 跨lncRNA基因座转录具有功能作用有学者指出,转录或剪接lncRNA的过程会影响和它在空间上最接近基因的表达,而与该lncRNA无关[64]。Engreitz等[65]认为,lncRNA基因Blustr的转录和剪接能够使得邻近的Sfmbt2表达升高,并且这一现象是由于转录剪接过程中所招募的多聚酶、染色体修饰酶作用的关系,而不受Blustr序列的影响。这一现象是不是影响基因表达的普遍作用模式尚待进一步研究。
3.8 LncRNA基因座可能含有影响邻近基因的功能元件有研究发现,删除lncRNA Bendr基因座的启动子区会显著降低邻近蛋白编码基因Bend4的转录和表达;但是在Bendr的第1个内含子中插入多聚腺苷酸信号,使得Bendr过早地终止转录,却并不影响Bend4的表达[65]。对lincRNA-p21功能的深入研究也发现了类似现象:即使在无法检测到lincRNA-p21表达的野生型小鼠组织中,敲除lincRNA-p21基因座同样会显著影响邻近基因Cdkn1a的表达[66]。以上研究提示lncRNA可能存在一种全新的调控机制:lncRNA基因座通过其中含有的特殊功能元件,比如增强子,发挥对邻近基因的顺式调控作用,而与lncRNA本身的序列和是否转录无关。这一假说的更多细节和普适性也需要更深入地研究。
4 天然免疫相关非编码RNA的研究策略在天然免疫领域,得益于近年来诸多新实验技术体系的出现与完善,非编码RNA相关的研究正日趋深入,未来进一步探索功能性非编码RNA及其作用机制仍将是主要的研究方向。目前,其在功能和机制方面的探索主要采用2个策略:(1)以非编码RNA为对象入手;(2)从某个分子作用模式入手。
4.1 以非编码RNA为对象入手从不同刺激或处理的转录组或表达组入手,寻找差异表达的非编码RNA。通过差异倍数和显著性及非编码RNA的基因组定位信息等筛选功能性候选RNA分子。然后通过后续干扰siRNA或Cas9[58, 67]进行功能筛选,锁定功能比较显著的RNA分子。最后进行RNA功能的分子机制研究,包括确定其相互作用分子、作用结合位点和作用的分子机制等。主要用到的技术有RNA-pulldown[68]、RIP[36]、ChIRP[69]等蛋白与RNA、DNA与RNA相互作用实验技术[10]。该策略是非编码RNA研究的常规套路,稳定可靠、风险较小,而后期分子机制研究一般是课题进展的难点,比较适用于大规模临床样品的课题和独特实验模型的课题。
4.2 从某个分子作用模式入手与第1种策略相反,该策略先靶向一个重要的蛋白分子[70],可以是信号转导分子、酶类或者转录因子等,或者是一个细胞亚结构,如线粒体、核内旁斑[71]、外泌体等,通过RIP-seq[72]或者RNA-seq检测其结合的或者包含的RNA,按照富集的倍数和显著性筛选候选RNA分子,之后通过siRNA或者高表达的方法筛选功能RNA[73]。该策略的优点是从课题设计开始就有明确指向的功能分子机制,在后续的分子机制研究中比较方便展开;而难点是前期如何做好RIP-seq和细胞亚结构的有效分离,这是后续实验可靠性和可行性的重要保障。
2种策略在实验技术上有部分重叠,但也有各自独特的实验技术需求或数据分析策略。不同策略适应于不同的课题和实验室背景,在选择时可以根据课题特点和实验室技术体系进行取舍。当然,2种策略也可以同时应用,相得益彰,相互印证,获得更好验证效果。
5 小结LncRNA在调控免疫细胞分化发育、调节炎性细胞因子的产生以及参与病毒和宿主的相互作用等天然免疫过程中具有重要的调控作用。目前lncRNA的研究方向主要集中在探索不同lncRNA的功能及其分子机制,近年来随着高通量测序和芯片技术的广泛应用,生物信息学算法的进步以及诸如RNA-pulldown、RIP、ChIRP、Crispr基因编辑、单细胞测序等新实验技术体系的出现与完善,筛选lncRNA的成功率与效率都不断提高,发现并注释了大量的lncRNA,出现了许多相对完善或别具特色的lncRNA数据库,例如NONCODE、GENCODE、lncRNAdb、CHIPbase、lncRNome、Starbase、lncRNADisease、lncATLAS等,为lncRNA生物功能的预测奠定了基础。同时,其他一些研究方向如lncRNA的可变剪接、lncRNA在转录后的编辑、lncRNA的表观修饰、lncRNA的高级结构和生物功能预测、lncRNA的转运、lncRNA在细胞自噬中的作用、lncRNA与外泌体的生成和转运等成为新的研究热点。我们相信随着研究的广泛开展与深入,lncRNA相关生物学效应及其分子机制将进一步得到阐明,并为疾病的诊断和治疗提供新的思路和靶点。
| [1] | LI L J, LENG R X, FAN Y G, PAN H F, YE D Q. Translation of noncoding RNAs:focus on lncRNAs, pri-miRNAs, and circRNAs[J]. Exp Cell Res, 2017, 361: 1–8. DOI: 10.1016/j.yexcr.2017.10.010 |
| [2] | NONCODE. http://www.noncode.org/analysis.php. |
| [3] | DJEBALI S, DAVIS C A, MERKEL A, DOBIN A, LASSMANN T, MORTAZAVI A, et al. Landscape of transcription in human cells[J]. Nature, 2012, 489: 101–108. DOI: 10.1038/nature11233 |
| [4] | LIU S J, HORLBECK M A, CHO S W, BIRK H S, MALATESTA M, HE D, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells[J/OL]. Science, 2017, 355. pii: aah7111. doi: 10.1126/science.aah7111. |
| [5] | lncATLAS. http://lncatlas.crg.eu/. |
| [6] | ZHAO Y, LI H, FANG S, KANG Y, WU W, HAO Y, et al. NONCODE 2016:an informative and valuable data source of long non-coding RNAs[J]. Nucleic Acids Res, 2016, 44(D1): D203–D208. DOI: 10.1093/nar/gkv1252 |
| [7] | GUTTMAN M, AMIT I, GARBER M, FRENCH C, LIN M F, FELDSER D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals[J]. Nature, 2009, 458: 223–227. DOI: 10.1038/nature07672 |
| [8] | LI Z, CHAO T C, CHANG K Y, LIN N, PATIL V S, SHIMIZU C, et al. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL[J]. Proc Natl Acad Sci USA, 2014, 111: 1002–1007. DOI: 10.1073/pnas.1313768111 |
| [9] | RAPICAVOLI N A, QU K, ZHANG J, MIKHAIL M, LABERGE R M, CHANG H Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics[J/OL]. Elife, 2013, 2: e00762. doi: 10.7554/eLife.00762. |
| [10] | WANG P, XUE Y, HAN Y, LIN L, WU C, XU S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation[J]. Science, 2014, 344: 310–313. DOI: 10.1126/science.1251456 |
| [11] | CARPENTER S, AIELLO D, ATIANAND M K, RICCI E P, GANDHI P, HALL L L, et al. A long noncoding RNA mediates both activation and repression of immune response genes[J]. Science, 2013, 341: 789–792. DOI: 10.1126/science.1240925 |
| [12] | PENG X, GRALINSKI L, ARMOUR C D, FERRIS M T, THOMAS M J, PROLL S, et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling[J/OL]. MBio, 2010, 1. pii: e00206-10. doi: 10.1128/mBio.00206-10. |
| [13] | DAVE R K, DINGER M E, ANDREW M, ASKARIAN-AMIRI M, HUME D A, KELLIE S. Regulated expression of PTPRJ/CD148 and an antisense long noncoding RNA in macrophages by proinflammatory stimuli[J/OL]. PLoS One, 2013, 8: e68306. doi: 10.1371/journal.pone.0068306. |
| [14] | GARMIRE L X, GARMIRE D G, HUANG W, YAO J, GLASS C K, SUBRAMANIAM S. A global clustering algorithm to identify long intergenic non-coding RNA-with applications in mouse macrophages[J/OL]. PLoS One, 2011, 6: e24051. doi: 10.1371/journal.pone.0024051. |
| [15] | YANG Y, WEN L, ZHU H. Unveiling the hidden function of long non-coding RNA by identifying its major partner-protein[J/OL]. Cell Biosci, 2015, 5: 59. doi: 10.1186/s13578-015-0050-x. |
| [16] | FATICA A, BOZZONI I. Long non-coding RNAs:new players in cell differentiation and development[J]. Nat Rev Genet, 2014, 15: 7–21. |
| [17] | BUNCH H. Gene regulation of mammalian long non-coding RNA[J]. Mol Genet Genomics, 2018, 293: 1–15. DOI: 10.1007/s00438-017-1370-9 |
| [18] | TAFT R J, PHEASANT M, MATTICK J S. The relationship between non-protein-coding DNA and eukaryotic complexity[J]. Bioessays, 2007, 29: 288–299. DOI: 10.1002/(ISSN)1521-1878 |
| [19] | ZHANG J, TAO Z, WANG Y. Long non-coding RNA DANCR regulates the proliferation and osteogenic differentiation of human bone-derived marrow mesenchymal stem cells via the p38 MAPK pathway[J]. Int J Mol Med, 2018, 41: 213–219. |
| [20] | JOLLY C, LAKHOTIA S C. Human sat Ⅲ and Drosophila hsr omega transcripts:a common paradigm for regulation of nuclear RNA processing in stressed cells[J]. Nucleic Acids Res, 2006, 34: 5508–5514. DOI: 10.1093/nar/gkl711 |
| [21] | YOON J H, ABDELMOHSEN K, SRIKANTAN S, YANG X, MARTINDALE J L, DE S, et al. LincRNA-p21 suppresses target mRNA translation[J]. Mol Cell, 2012, 47: 648–655. DOI: 10.1016/j.molcel.2012.06.027 |
| [22] | MAQUAT L E. Nonsense-mediated mRNA decay:splicing, translation and mRNP dynamics[J]. Nat Rev Mol Cell Biol, 2004, 5: 89–99. DOI: 10.1038/nrm1310 |
| [23] | WANG P, XU J, WANG Y, CAO X. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism[J]. Science, 2017, 358: 1051–1055. DOI: 10.1126/science.aao0409 |
| [24] | LEE J T. The X as model for RNA's niche in epigenomic regulation[J/OL]. Cold Spring Harb Perspect Biol, 2010, 2: a003749. doi: 10.1101/cshperspect.a003749. |
| [25] | TSAI M C, MANOR O, WAN Y, MOSAMMAPARAST N, WANG J K, LAN F, et al. Long noncoding RNA as modular scaffold of histone modification complexes[J]. Science, 2010, 329: 689–693. DOI: 10.1126/science.1192002 |
| [26] | BROSIUS J. Waste not, want not-transcript excess in multicellular eukaryotes[J]. Trends Genet, 2005, 21: 287–288. DOI: 10.1016/j.tig.2005.02.014 |
| [27] | IYER M K, NIKNAFS Y S, MALIK R, SINGHAL U, SAHU A, HOSONO Y, et al. The landscape of long noncoding RNAs in the human transcriptome[J]. Nat Genet, 2015, 47: 199–208. DOI: 10.1038/ng.3192 |
| [28] | LI R, ZHU H, LUO Y. Understanding the functions of long non-coding RNAs through their higher-order structures[J/OL]. Int J Mol Sci, 2016, 17. pii: E702. doi: 10.3390/ijms17050702. |
| [29] | KOTZIN J J, SPENCER S P, McCRIGHT S J, KUMAR D B U, COLLET M A, MOWEL W K, et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan[J]. Nature, 2016, 537: 239–243. DOI: 10.1038/nature19346 |
| [30] | ZHANG Y, CAO X. Long noncoding RNAs in innate immunity[J]. Cell Mol Immunol, 2016, 13: 138–147. DOI: 10.1038/cmi.2015.68 |
| [31] | MURPHY M B, MEDVEDEV A E. Long noncoding RNAs as regulators of Toll-like receptor signaling and innate immunity[J]. J Leukoc Biol, 2016, 99: 839–850. DOI: 10.1189/jlb.2RU1215-575R |
| [32] | AUNE T M, SPURLOCK C F 3rd. Long non-coding RNAs in innate and adaptive immunity[J]. Virus Res, 2016, 212: 146–160. DOI: 10.1016/j.virusres.2015.07.003 |
| [33] | CARPENTER S. Long noncoding RNA:novel links between gene expression and innate immunity[J]. Virus Res, 2016, 212: 137–145. DOI: 10.1016/j.virusres.2015.08.019 |
| [34] | CHEN M T, LIN H S, SHEN C, MA Y N, WANG F, ZHAO H L, et al. PU.1-regulated long noncoding RNA lnc-MC controls human monocyte/macrophage differentiation through interaction with microRNA 199a-5p[J]. Mol Cell Biol, 2015, 35: 3212–3224. |
| [35] | MOWEL W K, McCRIGHT S J, KOTZIN J J, COLLET M A, UYAR A, CHEN X, et al. Group 1 innate lymphoid cell lineage identity is determined by a cis-regulatory element marked by a long non-coding RNA[J/OL]. Immunity, 2017, 47: 435-449. e8. doi: 10.1016/j.immuni.2017.08.012. |
| [36] | LIU B, YE B, YANG L, ZHU X, HUANG G, ZHU P, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression[J]. Nat Immunol, 2017, 18: 499–508. DOI: 10.1038/ni.3712 |
| [37] | KRAWCZYK M, EMERSON B M. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes[J/OL]. Elife, 2014, 3: e01776. doi: 10.7554/eLife.01776. |
| [38] | IMAMURA K, IMAMACHI N, AKIZUKI G, KUMAKURA M, KAWAGUCHI A, NAGATA K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli[J]. Mol Cell, 2014, 53: 393–406. DOI: 10.1016/j.molcel.2014.01.009 |
| [39] | CHAN J, ATIANAND M, JIANG Z, CARPENTER S, AIELLO D, ELLING R, et al. Cutting edge:a natural antisense transcript, AS-IL1α, controls inducible transcription of the proinflammatory cytokine IL-1α[J]. J Immunol, 2015, 195: 1359–1363. DOI: 10.4049/jimmunol.1500264 |
| [40] | KAMBARA H, NIAZI F, KOSTADINOVA L, MOONKA D K, SIEGEL C T, POST A B, et al. Negative regulation of the interferon response by an interferon-induced long non-coding RNA[J]. Nucleic Acids Res, 2014, 42: 10668–10680. DOI: 10.1093/nar/gku713 |
| [41] | LU J, WU X, HONG M, TOBIAS P, HAN J. A potential suppressive effect of natural antisense IL-1β RNA on lipopolysaccharide-induced IL-1β expression[J]. J Immunol, 2013, 190: 6570–6578. DOI: 10.4049/jimmunol.1102487 |
| [42] | TYCOWSKI K T, GUO Y E, LEE N, MOSS W N, VALLERY T K, XIE M, et al. Viral noncoding RNAs:more surprises[J]. Genes Dev, 2015, 29: 567–584. DOI: 10.1101/gad.259077.115 |
| [43] | GOMEZ J A, WAPINSKI O L, YANG Y W, BUREAU J F, GOPINATH S, MONACK D M, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus[J]. Cell, 2013, 152: 743–754. DOI: 10.1016/j.cell.2013.01.015 |
| [44] | AMIT-AVRAHAM I, POZNER G, ESHAR S, FASTMAN Y, KOLEVZON N, YAVIN E, et al. Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum[J/OL]. Proc Natl Acad Sci USA, 2015, 112: E982-E991. doi: 10.1073/pnas.1420855112. |
| [45] | COLLIER S P, COLLINS P L, WILLIAMS C L, BOOTHBY M R, AUNE T M. Cutting edge:influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells[J]. J Immunol, 2012, 189: 2084–2088. DOI: 10.4049/jimmunol.1200774 |
| [46] | ZHANG Q, CHEN C Y, YEDAVALLI V S, JEANG K T. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression[J/OL]. MBio, 2013, 4: e00596-12. doi: 10.1128/mBio.00596-12. |
| [47] | LIU Y, PAN S, LIU L, ZHAI X, LIU J, WEN J, et al. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population[J/OL]. PLoS One, 2012, 7: e35145. doi: 10.1371/journal.pone.0035145. |
| [48] | WANG J, LIU X, WU H, NI P, GU Z, QIAO Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer[J]. Nucleic Acids Res, 2010, 38: 5366–5383. DOI: 10.1093/nar/gkq285 |
| [49] | IWAKIRI D, ZHOU L, SAMANTA M, MATSUMOTO M, EBIHARA T, SEYA T, et al. Epstein-Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3[J]. J Exp Med, 2009, 206: 2091–2099. DOI: 10.1084/jem.20081761 |
| [50] | LI Y, WANG C, MIAO Z, BI X, WU D, JIN N, et al. ViRBase:a resource for virus-host ncRNA-associated interactions[J]. Nucleic Acids Res, 2015, 43(Database issue): D578–D582. |
| [51] | CAMPBELL M, KUNG H J, IZUMIYA Y. Long non-coding RNA and epigenetic gene regulation of KSHV[J]. Viruses, 2014, 6: 4165–4177. DOI: 10.3390/v6114165 |
| [52] | ROSSETTO C C, PARI G. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome[J/OL]. PLoS Pathog, 2012, 8: e1002680. doi: 10.1371/journal.ppat.1002680. |
| [53] | LUDWIG L B, AMBRUS J L Jr, KRAWCZYK K A, SHARMA S, BROOKS S, HSIAO C B, et al. Human immunodeficiency virus-type 1 LTR DNA contains an intrinsic gene producing antisense RNA and protein products[J/OL]. Retrovirology, 2006, 3: 80. doi: 10.1186/1742-4690-3-80. |
| [54] | LANDRY S, HALIN M, LEFORT S, AUDET B, VAQUERO C, MESNARD J M, et al. Detection, characterization and regulation of antisense transcripts in HIV-1[J]. Retrovirology, 2007, 4: 71. DOI: 10.1186/1742-4690-4-71 |
| [55] | KOBAYASHI-ISHIHARA M, YAMAGISHI M, HARA T, MATSUDA Y, TAKAHASHI R, MIYAKE A, et al. HIV-1-encoded antisense RNA suppresses viral replication for a prolonged period[J]. Retrovirology, 2012, 9: 38. DOI: 10.1186/1742-4690-9-38 |
| [56] | SAAYMAN S, ACKLEY A, TURNER A W, FAMIGLIETTI M, BOSQUE A, CLEMSON M, et al. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription[J]. Mol Ther, 2014, 22: 1164–1175. DOI: 10.1038/mt.2014.29 |
| [57] | CLEMSON C M, HUTCHINSON J N, SARA S A, ENSMINGER A W, FOX A H, CHESS A, et al. An architectural role for a nuclear noncoding RNA:NEAT1 RNA is essential for the structure of paraspeckles[J]. Mol Cell, 2009, 33: 717–726. DOI: 10.1016/j.molcel.2009.01.026 |
| [58] | HUANG W, THOMAS B, FLYNN R A, GAVZY S J, WU L, KIM S V, et al. DDX5 and its associated lncRNA RMRP modulate TH17 cell effector functions[J]. Nature, 2015, 528: 517–522. DOI: 10.1038/nature16193 |
| [59] | HUARTE M, GUTTMAN M, FELDSER D, GARBER M, KOZIOL M J, KENZELMANN-BROZ D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response[J]. Cell, 2010, 142: 409–419. DOI: 10.1016/j.cell.2010.06.040 |
| [60] | WILUSZ J E, FREIER S M, SPECTOR D L. 3'end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA[J]. Cell, 2008, 135: 919–932. DOI: 10.1016/j.cell.2008.10.012 |
| [61] | TRIPATHI V, ELLIS J D, SHEN Z, SONG D Y, PAN Q, WATT A T, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation[J]. Mol Cell, 2010, 39: 925–938. DOI: 10.1016/j.molcel.2010.08.011 |
| [62] | KIM Y K, FURIC L, DESGROSEILLERS L, MAQUAT L E. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay[J]. Cell, 2005, 120: 195–208. DOI: 10.1016/j.cell.2004.11.050 |
| [63] | GONG C, MAQUAT L E. LncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3'UTRs via Alu elements[J]. Nature, 2011, 470: 284–288. DOI: 10.1038/nature09701 |
| [64] | EBISUYA M, YAMAMOTO T, NAKAJIMA M, NISHIDA E. Ripples from neighbouring transcription[J]. Nat Cell Biol, 2008, 10: 1106–1113. DOI: 10.1038/ncb1771 |
| [65] | ENGREITZ J M, HAINES J E, PEREZ E M, MUNSON G, CHEN J, KANE M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing[J]. Nature, 2016, 539: 452–455. DOI: 10.1038/nature20149 |
| [66] | GROFF A F, SANCHEZ-GOMEZ D B, SORUCO M M L, GERHARDINGER C, BARUTCU A R, LI E, et al. In vivo characterization of linc-p21 reveals functional cis-regulatory DNA elements[J]. Cell Rep, 2016, 16: 2178–2186. DOI: 10.1016/j.celrep.2016.07.050 |
| [67] | RAN F A, CONG L, YAN W X, SCOTT D A, GOOTENBERG J S, KRIZ A J, et al. In vivo genome editing using Staphylococcus aureus Cas9[J]. Nature, 2015, 520: 186–191. DOI: 10.1038/nature14299 |
| [68] | ATIANAND M K, HU W, SATPATHY A T, SHEN Y, RICCI E P, ALVAREZ-DOMINGUEZ J R, et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation[J]. Cell, 2016, 165: 1672–1685. DOI: 10.1016/j.cell.2016.05.075 |
| [69] | CHU C, QUINN J, CHANG H Y. Chromatin isolation by RNA purification (ChIRP)[J/OL]. J Vis Exp, 2012(61). pii: 3912. doi: 10.3791/3912. |
| [70] | LIEPELT A, NAARMANN-DE VRIES I S, SIMONS N, EICHELBAUM K, FÖHR S, ARCHER S K, et al. Identification of RNA-binding proteins in macrophages by interactome capture[J]. Mol Cell Proteomics, 2016, 15: 2699–2714. DOI: 10.1074/mcp.M115.056564 |
| [71] | ADRIAENS C, STANDAERT L, BARRA J, LATIL M, VERFAILLIE A, KALEV P, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity[J]. Nat Med, 2016, 22: 861–868. DOI: 10.1038/nm.4135 |
| [72] | VAN NOSTRAND E L, PRATT G A, SHISHKIN A A, GELBOIN-BURKHART C, FANG M Y, SUNDARARAMAN B, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP)[J]. Nat Methods, 2016, 13: 508–514. DOI: 10.1038/nmeth.3810 |
| [73] | XING Y H, YAO R W, ZHANG Y, GUO C J, JIANG S, XU G, et al. SLERT regulates DDX21 rings associated with PolⅠtranscription[J/OL]. Cell, 2017, 169: 664-678. e16. doi: 10.1016/j.cell.2017.04.011. |
 2018, Vol. 39
2018, Vol. 39


