2. 第二军医大学基础部医学遗传学研究所, 上海 200433
2. Institute of Medical Genetics, College of Basic Medical Sciences, Second Military Medical University, Shanghai 200433, China
尿石症(urolithiasis)的发病率呈逐年上升趋势,正逐渐成为全球性的公共卫生问题[1]。结晶是肾结石的早期表现形式,其在肾脏沉积引起肾结石是尿石症的主要发病机制。尽管目前对于肾结晶形成的具体发病机制尚未完全阐明,但已有相当多的证据表明结晶在肾脏的沉积与氧化应激、炎症反应等密切相关[2-3]。其中,肾小管上皮受到结晶刺激后分泌骨桥蛋白(osteopontin,OPN),再协同其他炎症介质一起募集巨噬细胞至病变部位,这是贯穿结晶肾损伤的核心环节[3]。
近年来长链非编码RNA (long non-coding RNA,lncRNA)调控多种疾病的发病正逐渐成为各领域的研究热点,目前最新lncRNA的鉴定及功能研究主要集中在肿瘤转移、神经退行性疾病以及自身免疫性疾病等方面[4-6],而在肾脏病领域,相关研究目前尚处于起步阶段,仅有少数lncRNA差异表达在肾脏疾病中有相关性研究报道[7-8]。本研究通过构建草酸钠诱导的肾小管上皮细胞结晶肾损伤细胞模型,初步探讨OPN相关lncRNA在结晶肾损伤中的调控作用,进一步拓展对肾结晶发生、发展的认识。
1 材料和方法 1.1 细胞培养人肾近曲小管上皮细胞HK-2(货号SCSP-511) 购自中国科学院上海细胞培养中心。细胞贴壁生长于含10%胎牛血清、100 U/mL青霉素、100 mg/mL链霉素的DMEM F/12培养液,置于37 ℃、5% CO2培养箱培养。第7~9代细胞用于实验。
1.2 小干扰RNA(siRNA)转染与草酸钠结晶肾损伤细胞模型构建将细胞随机分为2组:对照组(NC-siRNA组)和干扰组(OPN-siRNA组),NC-siRNA组细胞转染阴性随机对照序列,OPN-siRNA组细胞转染OPN siRNA,两组细胞转染后均给予草酸钠(20 mmol/L[9];上海将来实业有限公司)处理细胞12 h构建草酸钠结晶肾损伤细胞模型。每组实验重复3次。本研究采用的siRNA由美国Invitrogen公司合成,转染试剂为美国Invitrogen公司研发的LipofectamineTM 3000细胞脂质体转染试剂盒,将该脂质体、siRNA、OPTI-MEM培养液(美国Gibco公司)充分混匀,混合物室温静置5 min后加入含细胞的培养皿中,转染48 h后收取细胞。
1.3 LncRNA芯片检测按照TRIzol试剂盒(美国Invitrogen公司)操作说明提取细胞总RNA;用超微量紫外分光光度计(NanoDrop 2000) 检测波长260 nm和280 nm处的光密度(D)值并定量;用RNA甲醛变性凝胶电泳分析RNA质量。采用美国Arraystar公司提供的人类lncRNA芯片(V4.0)、根据美国Agilent Technologies公司的彩色微阵列的基因表达分析(One-Color Microarray-Based Gene Expression Analysis)实验方案检测40 173个lncRNA和20 730个mRNA的表达。
1.4 差异lncRNA与mRNA的生物信息学分析对lncRNA芯片数据进行标准化处理和组间比较后筛选差异表达的lncRNA和mRNA,筛选标准:差异倍数(fold change)>1.5且P<0.05。采用limma算法对两组差异表达的lncRNA与mRNA进行分析和计算。使用GeneCodis软件编写脚本程序,对lncRNA芯片数据进行关联分析和层次聚类分析;运用基因本体论(gene ontology,GO)和京都基因与基因组百科全书数据库(Kyoto encyclopedia of genes and genomes,KEGG)对两组差异表达的lncRNA与mRNA进行功能和生物学行为分析。
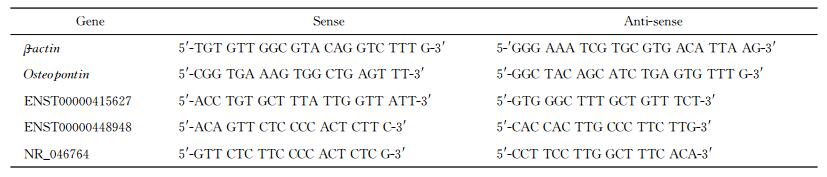
1.5 qPCR验证细胞转染和差异表达用qPCR验证OPN-siRNA转染后细胞OPN mRNA的表达以及对lncRNA芯片筛选出的差异倍数最高的3个lncRNA(ENST00000448948、ENST00000415627和NR_046764) 进行验证。提取细胞总RNA并定量后,用第1链cDNA合成试剂盒(货号C28025-011,美国Invitrogen公司)按操作说明合成cDNA。采用日本TaKaRa公司的SYBR预混酶,以cDNA为模板、β-actin作为内参照基因,运用美国Applied Biosystems公司的实时定量PCR检测系统(StepOneTM Real-Time PCR System)进行qPCR反应。用Primer Premier 6.0设计引物,引物序列见表 1。反应体系为20 μL,含10 μL SYBR Green、1 μalign="left"引物、1 μL cDNA。循环条件:95 ℃ 10 min;95 ℃ 15 s、60 ℃ 15 s、72 ℃ 20 s,40个循环。实验重复3次,并根据实际情况设置复孔,并设置相应的无模板对照组。
|
|
表 1 引物序列 Tab 1 Primer sequences |
1.6 统计学处理
应用SPSS 17.0软件进行统计学分析,计量资料用x±s表示,非正态分布资料采用非参数方法(Kruskal-Wallis)秩和检验,两组间比较采用Mann-Whitney U检验。检验水准(α)为0.05。
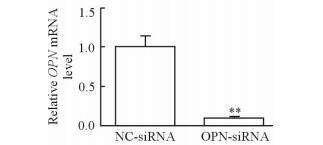
2 结果 2.1 OPN-siRNA细胞构建的验证通过qPCR检测OPN-siRNA组和NC-siRNA组细胞中OPN mRNA的表达,结果(图 1)显示OPN-siRNA组OPN mRNA表达降低(P<0.01),表明OPN干扰效果较好,OPN-siRNA细胞成功构建。
|
图 1 qPCR验证OPN-siRNA细胞构建成功 Fig 1 Expression of OPN mRNA by qPCR NC-siRNA group: The cells were transfected with negative random control sequence; OPN-siRNA group: The cells were transfected with OPN siRNA. OPN: Osteopontin.**P < 0.01 vs NC-siRNA group. n=3, x±s |
2.2 OPN相关lncRNA与mRNA的差异表达
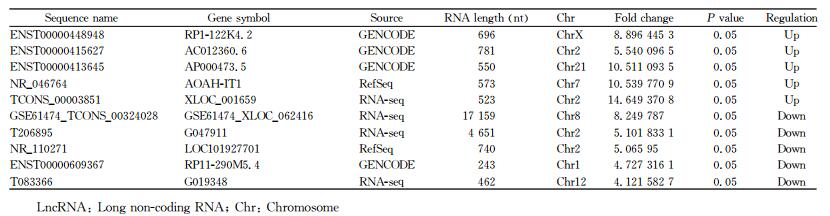
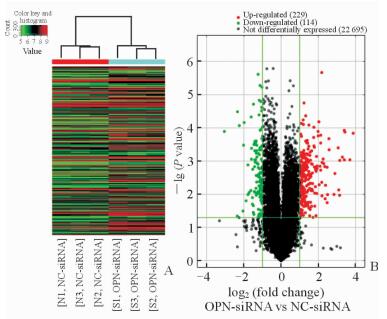
运用Arraystar人类lncRNA芯片(V4.0) 检测40 173个lncRNA和20 730个mRNA,筛选得到差异表达的lncRNA和mRNA。采用聚类分析热图(图 2A)发现差异表达lncRNA共583个,其中OPN-siRNA组表达上调354个、下调229个;差异表达mRNA共235个,其中OPN-siRNA组表达上调139个、下调96个。火山图(图 2B)红色区域表示差异表达lncRNA。对差异表达lncRNA进行数据筛选,筛选出OPN-siRNA组与NC-siRNA组相比差异表达上调和下调差异倍数分别位于前5位的lncRNA(表 2)。
|
图 2 LncRNA聚类分析热图(A)和火山图(B) Fig 2 LncRNA clustering analysis plot (A) and volcano plot (B) Red color indicates up-regulated expression and green color indicates down-regulated expression in Fig 2A. NC-siRNA group: The cells were transfected with negative random control sequence; OPN-siRNA group: The cells were transfected with OPN siRNA. LncRNA: Long non-coding RNA; OPN: Osteopontin |
|
|
表 2 10个差异表达倍数最高的lncRNA Tab 2 Ten lncRNAs with the greatest expression difference |
2.3 备选lncRNA表达的验证
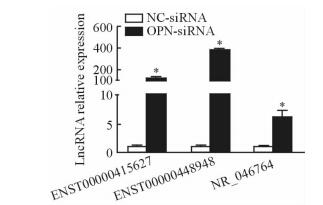
通过qPCR验证筛选出的差异倍数最高的3个lncRNA:ENST00000448948(fold change=8.896 445 3)、ENST00000415627(fold change=5.540 096 5)、NR_046764(fold change=10.539 770 9) 进行验证,其在OPN-siRNA组表达均高于NC-siRNA组(P<0.05,图 3)。
|
图 3 qPCR验证差异表达lncRNA Fig 3 Differentially expressed lncRNAs by qPCR NC-siRNA group: The cells were transfected with negative random control sequence; OPN-siRNA group: The cells were transfected with OPN siRNA. LncRNA: Long non-coding RNA; OPN: Osteopontin.*P < 0.05 vs NC-siRNA group. n=3, x±s |
2.4 差异表达lncRNA生物信息学分析
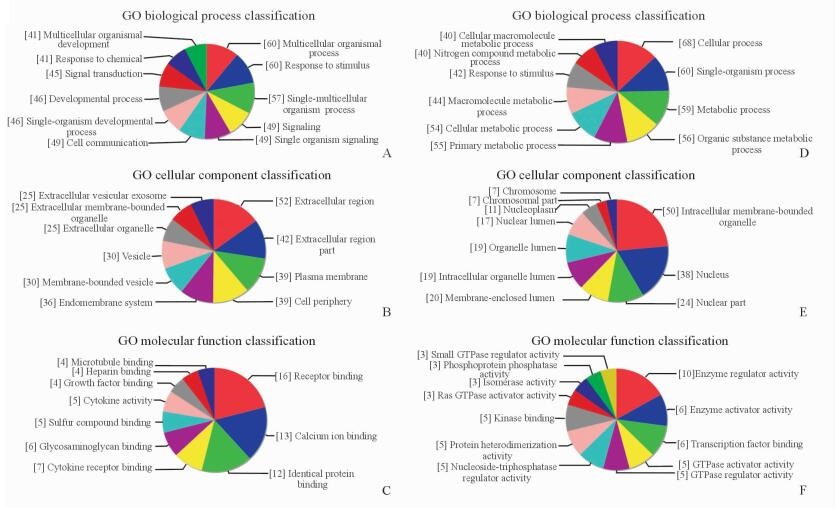
通过对差异lncRNA与其邻近相关mRNA进行联合分析可知,差异表达的lncRNA主要参与细胞内信号转导、细胞增殖、细胞凋亡、炎症反应等生物学过程。图 4所示为差异表达lncRNA的GO分析结果。其中上调转录本靶向作用最显著的GO是细胞外基质形成(GO:0030198;图 4A)、细胞外结构域(GO:0005509;图 4B)、细胞外受体结合位点(GO:0005576;图 4C),而下调转录本靶向作用最显著的GO是细胞进程(GO:0009987;图 4D)、细胞器膜结合位点(GO:0005667;图 4E)、酶活性调节(GO:0030234;图 4F)。
|
图 4 GO分析差异表达的lncRNA Fig 4 Gene ontology (GO) analysis of differentially expressed lncRNAs The top several GOs were showed in the most significant enrichment (fold change > 2, P < 0.05). A-C: The up-regulated GO targets; D-F: The down-regulated GO targets. LncRNA: Long non-coding RNA |
3 讨论
肾结石是临床常见病、多发病,多见于20~60岁人群,且复发率较高[10]。尽管目前治疗肾结石方法较多,但其高复发率使得肾结石仍严重影响患者的生活质量,因此,扩展肾结石的研究方向,探寻有效治疗和预防肾结石复发的靶点十分重要。目前研究发现代谢异常与尿石症存在共生和伴随关系[11],但其具体机制仍不清楚。探讨肾结石的早期病变——肾结晶,及其诱导的肾损伤以及修复机制是近年来该领域的研究热点。
LncRNA作为真核生物基因组组成部分之一,在多种疾病的发病机制中具有重要作用。尽管大部分lncRNA的功能仍然未知,但作为一种重要的调控真核细胞转录的分子已被广泛报道,可以在表观遗传、转录调控以及转录后调控等多个层面影响基因的表达[12]。研究发现lncRNA在多种疾病中对编码蛋白基因具有调控作用[13]。Yang等[14]研究发现lncRNA-LET表达下调参与了低氧诱导的肝癌侵袭转移过程。Lorenzen等[15]研究发现循环血中lncRNA-TapSAKI的表达量可能是预测急性肾损伤(AKI)患者死亡的指标之一,而肾结石相关lncRNA的研究更是少之又少。
OPN是肾小管上皮细胞分泌的大分子物质,且是肾结石有机基质的组成部分[16],在肾结石形成过程中具有关键作用。既往研究表明OPN可通过精氨酸-甘氨酸-天冬氨酸序列(RGD)以及钙结合位点结合钙离子,从而促进含钙结石形成[17];体外培养犬肾细胞(MDCK细胞)中观察到OPN可以促进草酸钙结石黏附[18]。此外,OPN作为一种细胞外基质蛋白能通过抑制转录因子IRF1激活lncRNA HOTAIR的表达,使肿瘤细胞的侵袭性增加、恶性化程度增高[19]。鉴于OPN在肾脏疾病中的重要作用以及前期研究揭示的结晶肾损伤小鼠模型中lncRNA差异表达[20],我们推测lncRNA可能参与调控OPN在结晶肾损伤中的作用。
本研究首次利用生物芯片技术及生物信息学方法对结晶肾损伤中OPN相关lncRNA以及mRNA进行了联合分析,确定OPN-siRNA组有583个lncRNA差异表达。进一步运用生物信息学方法对潜在的靶基因进行GO和KEGG分析:GO分析结果表明差异表达lncRNA与细胞分化、细胞外基质形成、酶活性调节等相关联;KEGG分析结果发现差异表达lncRNA对应多条细胞内信号通路。其中NOD样受体信号通路、AGE-RAGE信号通路、炎症性肠病相关信号通路、细胞周期调控、脂肪酸的生物合成以及TGF-β受体信号通路得到显著富集。
LncRNA常通过微小RNA(miRNA)介导的内源性竞争性RNA以及miRNA“海绵吸附”作用机制来调控各种生物学进程[21]。1个lncRNA分子可以吸附多个miRNA,而1个miRNA分子可调节多个靶基因[21-22]。有研究表明miR-21可以通过调控PI3K/AKT/mTOR信号通路对TGF-β1基因进行调控[23],lncRNA H19可以在胎盘发育的后期表达增加,并通过与miR-675相互作用抑制胎盘发育过度的作用[24]。因此,基于本研究lncRNA的生物信息学分析结果,我们后续研究将进一步探讨lncRNA调控OPN相关基因表达的不同机制,并猜测lncRNA也可能会通过miRNA分子参与OPN的调控作用,或与PI3K/AKT/mTOR信号以及TGF-β相关通路相互关联,从而参与结晶肾损伤的发生。
综上所述,本研究初步通过生物信息学方法,从GO和KEGG分析层面对OPN相关lncRNA可能参与的结晶肾损伤的发生机制进行初步探讨。随着对分子生物学以及表观遗传学研究的深入,本研究从基因分析角度探讨结晶肾损伤的发病机制,有助于阐明lncRNA在调控结晶肾损伤方面的功能和在分子机制中的作用,以期为该类疾病的防治提供线索和潜在干预靶点。
| [1] | CUNNINGHAM P, NOBLE H, Al-MODHEFER A K, WALSH I. Kidney stones:pathophysiology, diagnosis and management[J]. Br J Nurs, 2016, 25: 1112–1116. DOI: 10.12968/bjon.2016.25.20.1112 |
| [2] | HUANG H S, MA M C, CHEN J. Low-vitamin E diet exacerbates calcium oxalate crystal formation via enhanced oxidative stress in rat hyperoxaluric kidney[J]. Am J Physiol Renal Physiol, 2009, 296: F34–F45. |
| [3] | OKADA A, YASUI T, FUJⅡ Y, NⅡMI K, HAMAMOTO S, HIROSE M, et al. Renal macrophage migration and crystal phagocytosis via inflammatory-related gene expression during kidney stone formation and elimination in mice:detection by association analysis of stone-related gene expression and microstructural observation[J]. J Bone Miner Res, 2010, 25: 2425–2435. |
| [4] | NAGANO T, FRASER P. No-nonsense functions for long noncoding RNAs[J]. Cell, 2011, 145: 178–181. DOI: 10.1016/j.cell.2011.03.014 |
| [5] | CESANA M, CACCHIARELLI D, LEGNINI I, SANTINI T, STHANDIER O, CHINAPPI M, et al. A long noncoding RNA controls muscle differentiation by functioning as acompeting endogenous RNA[J]. Cell, 2011, 147: 358–369. DOI: 10.1016/j.cell.2011.09.028 |
| [6] | GEISLER S, LOJEK L, KHALIL A M, BAKER K E, COLLER J. Decapping of long noncoding RNAs regulates inducible genes[J]. Mol Cell, 2012, 45: 279–291. DOI: 10.1016/j.molcel.2011.11.025 |
| [7] | ZHOU Q, CHUNG A C, HUANG X R, DONG Y, YU X, LAN H Y. Identification of novel long noncoding RNAs associated with TGF-β/Smad3-mediated renal inflammation and fibrosis by RNA sequencing[J]. Am J Pathol, 2014, 184: 409–417. DOI: 10.1016/j.ajpath.2013.10.007 |
| [8] | SUI W, LI H, OU M, TANG D, DAI Y. Altered long non-coding RNA expression profile in patients with IgA-negative mesangial proliferative glomerulonephritis[J]. Int J Mol Med, 2012, 30: 173–178. |
| [9] | MARONI P D, KOUL S, MEACHAM R B, CHANDHOKE P S, KOUL H K. Effects of oxalate on IMCD cells:a line of mouse inner medullary collecting duct cells[J]. Ann N Y Acad Sci, 2004, 1030: 144–149. DOI: 10.1196/annals.1329.018 |
| [10] | MELLO M F, MARCHINI G S, CẬMARA C, DANILOVIC A, LEVY R, ELUF-NETO J, et al. A large 15-year database analysis on the influence of age, gender, race, obesity and income on hospitalization rates due to stone disease[J]. Int Braz J Urol, 2016, 42: 1150–1159. DOI: 10.1590/s1677-5538.ibju.2015.0743 |
| [11] | KHAN S R. Is oxidative stress, a link between nephrolithiasis and obesity, hypertension, diabetes, chronic kidney disease, metabolic syndrome?[J]. Urol Res, 2012, 40: 95–112. DOI: 10.1007/s00240-011-0448-9 |
| [12] | YI H, PENG R, ZHANG L Y, SUN Y, PENG H M, LIU H D, et al. LincRNA-Gm4419 knockdown ameliorates NF-κB/NLRP3 inflammasome-mediated inflammation in diabetic nephropathy[J/OL]. Cell Death Dis, 2017, 8:e2583. doi:10.1038/cddis.2016.451. http://europepmc.org/abstract/MED/28151474 |
| [13] | WAPINSKI O, CHANG H Y. Long noncoding RNAs and human disease[J]. Trends Cell Biol, 2011, 21: 354–361. DOI: 10.1016/j.tcb.2011.04.001 |
| [14] | YANG F, HUO X S, YUAN S X, ZHANG L, ZHOU W P, WANG F, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis[J]. Mol Cell, 2013, 49: 1083–1096. DOI: 10.1016/j.molcel.2013.01.010 |
| [15] | LORENZEN J M, SCHAUERTE C, KIELSTEIN J T, HUBNER A, MARTINO F, FIEDLER J, et al. Circulating long noncoding RNA TapSAKI is a predictor of mortality in citically ill patients with acute kidney injury[J]. Clin Chem, 2015, 61: 191–201. DOI: 10.1373/clinchem.2014.230359 |
| [16] | MILLER N L, EVAN A P, LINGEMAN J E. Pathogenesis of renal calculi[J]. Urol Clin North Am, 2007, 34: 295–313. DOI: 10.1016/j.ucl.2007.05.007 |
| [17] | HAMAMOTO S, NOMURA S, YASUI T, OKADA A, HIROSE M, SHIMIZU H, et al. Effects of impaired functional domains of osteopontin on renal crystal formation:analyses of OPN transgenic and OPN knockout mice[J]. J Bone Miner Res, 2010, 25: 2712–2723. |
| [18] | YAMATE T, KOHRI K, UMEKAWA T, AMASAKI N, ISIKAWA Y, IGUCHI M, et al. The effect of osteopontin on the adhesion of calcium oxalate crystals to Madin-Darby canine kidney cells[J]. Eur Urol, 1996, 30: 388–393. |
| [19] | YANG G, ZHANG S, GAO F, LIU Z, LU M J, PENG S, et al. Osteopontin enhances the expression of HOTAIR in cancer cells via IRF1[J]. Biochim Biophys Acta, 2014, 1839: 837–848. DOI: 10.1016/j.bbagrm.2014.06.020 |
| [20] | 张彩虹, 谌卫, 袁继行, 张杰, 马文杰, 刘敏, 等. 应用微阵列芯片分析草酸钙结晶肾损伤中LncRNA表达谱的差异[J]. 中国中西医结合肾病杂志, 2016, 17: 14–17. |
| [21] | TAY Y, RINN J, PANDOLFI P P. The multilayered complexity of ceRNA crosstalk and competition[J]. Nature, 2014, 505: 344–352. DOI: 10.1038/nature12986 |
| [22] | MUKHERJI S, EBERT M S, ZHENG G X, TSANG J S, SHARP P A, VAN OUDENAARDEN A. MicroRNAs can generate thresholds in target gene expression[J]. Nat Genet, 2011, 43: 854–859. DOI: 10.1038/ng.905 |
| [23] | BAI L, LIANG R, YANG Y, HOU X, WANG Z, ZHU S, et al. MicroRNA-21 regulates PI3K/AKT/mTOR signaling by targeting TGF βⅠ during skeletal muscle development in pigs[J/OL]. PLoS One, 2015, 10:e0119396. doi:10.1371/journal.pone.0119396. |
| [24] | KENIRY A, OXLEY D, MONNIER P, KYBA M, DANDOLO L, SMITS G, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r[J]. Nat Cell Biol, 2012, 14: 659–665. DOI: 10.1038/ncb2521 |
 2017, Vol. 38
2017, Vol. 38