自1953年DNA双螺旋结构被发现以来,遗传学领域取得了突飞猛进的发展。近年来,随着全基因组关联研究(genome wide association study,GWAS)、二代测序、三代测序等技术的问世,遗传学研究成为探索复杂性疾病致病机制的有力工具[1-3]。而高品质的DNA样本是遗传学实验成功与否的重要影响因素,进行有效的DNA样本质检更是控制实验成本的关键措施。以GWAS为例,各研究平台均需要200 ng的DNA,且要求DNA最低浓度为50 ng/μL[4]。NanoDrop和PicoGreen是检测DNA样本质量较为合适的方法,NanoDrop(如ND-1000)是一种全波长紫外/可见光扫描分光光度计,可检测DNA、RNA、蛋白质和染料等的光密度(D)值,自动识别的光谱范围为220~750 nm[5];PicoGreen为一种选择性结合双链DNA的荧光染料,作用与SYBR-GreenⅠ相似[6]。然而,研究获得的DNA等样本中存在杂质时,通过上述方法很难被检测出来,因此需要一种更加可靠的方法评估DNA样品的质量。本研究通过分析24例GWAS芯片失败样本在质量控制过程中的详细情况,发现失败样本的D260/D230值偏低,进而在更大样本中明确D260/D230值与DNA质量的关系,旨在寻找更可靠的DNA质检手段,降低芯片检测失败风险。
1 材料和方法 1.1 研究对象收集1 494例第二军医大学长征医院收治的强直性脊柱炎(ankylosing spondylitis, AS)患者的全血样本,AS诊断均符合1984年修订的AS美国纽约的诊断标准[7]。本研究通过第二军医大学长征医院医学伦理委员会审批,所有入组患者均签署知情同意书。使用AxyPrep血基因组DNA小量试剂盒提取所有患者血液样本DNA,用于后续检测。
1.2 NanoDrop检测检测样本前先对ND-1000(NanoDrop, USA)进行校正,取Lambda DNA原液(310 ng/μL,日本TaKaRa公司),用TE缓冲液稀释成3.1、38、78、155、310 ng/μL 5个浓度梯度,使用ND-1000检测各浓度样品在260 nm下的D值并绘制曲线,根据曲线拟合度判断仪器精确性。校正后取待测DNA样品1 μL,上机检测样品浓度及D280、D260、D230等值。
1.3 PicoGreen检测(1) DNA待测样品的准备:取1 μL DNA样品加入到349 μL的TE缓冲液中振荡混匀,另取3个空移液管,每管加入100 μL样品混匀液;(2)绘制标准曲线:取λDNA原液(310 ng/μL,日本TaKaRa公司)用TE缓冲液稀释至2 ng/μL,然后再稀释成0(不含DNA)、0.125、0.25、0.5、1 ng/μL 5个浓度梯度,检测并绘制标准曲线。(3)DNA样品检测:取DNA待测样品室温避光孵育5 min,应用FlexStation® 3酶标仪(Molecular Devices, USA)以480 nm为激发波长测定各样品在520 nm下的D值,并使用SoftMax® Pro软件进行分析。每组设3个复孔。
1.4 PCR检测管家基因选取NanoDrop和PicoGreen检测浓度均大于50 ng/μL的DNA样品行管家基因(GAPDH)的PCR检测。GAPDH引物(452 bp)序列:上游5′-ACC ACA GTC CAT GCC ATC AC-3′,下游5′-ATG TCG TTG TCC CAC CAC CT-3′。20 μL PCR体系:5 μL DNA模板(浓度10 ng/μL),10 μL Premix TAQ酶,上、下游引物各2 μL,1 μL去离子水。反应条件:95 ℃预变性10 min;94 ℃变性1 min、60 ℃退火1 min、72 ℃延伸1 min 25 s,共33个循环;循环结束后72 ℃延伸7 min。取10 μL PCR反应产物行带有GoldView染料(上海赛百盛基因技术有限公司)的琼脂糖凝胶电泳,验证是否出现452 bp的扩增条带。
1.5 基因分型采用Illumina公司的Human Omini ZhongHua-8 BeadChip芯片行基因分型检测,基因组DNA经过质控检测后,取合格样品稀释至浓度为50 ng/μL。第一阶段:选取24例NanoDrop和PicoGreen检测浓度>50 ng/μL的DNA样品行芯片检测,有8例芯片反应失败,比较16例成功样本与8例失败样本的NanoDrop检测的D260/D280值及D260/D230值。第二阶段:对NanoDrop和PicoGreen检测浓度>50 ng/μL且PCR-管家基因扩增成功的DNA样品行基因分型检测,并使用HD超微阵序列和iScan系统(Illumina公司)进行扫描。
1.6 统计学处理采用SPSS 13.0软件行统计分析。呈正态分布的计量资料以x±s表示,呈非正态分布的计量资料以中位数(最小值~最大值)表示。D260/D230值两组间比较采用Mann-Whitney U检验,采用受试者工作特征(receiver operating characteristic,ROC)曲线评价D260/D230值对PCR结果的鉴别效率,并确定最佳诊断点(敏感度和特异度最大)。为达到最严格的芯片质量控制,选取特异度为0.95的点为参考点;同时选取PicoGreen检测浓度>50 ng/μL的样本再计算ROC曲线以排除浓度对PCR结果的影响。使用曲线下面积(area under curve,AUC)评价D260/D230值对PCR结果的预测能力。检验水准(α)为0.05。
2 结果 2.1 NanoDrop与PicoGreen检测结果1 494例DNA样品的NanoDrop检测浓度为(131.3±116.2) ng/μL,D260/D280值为1.815±0.868,D260/D230值为1.37(0.08~18.61);PicoGreen检测浓度为(123.0±97.5) ng/μL。NanoDrop和PicoGreen检测浓度均大于50 ng/μL的样本有1 122例。
2.2 24例DNA样品芯片检测结果第一阶段24例NanoDrop和PicoGreen检测浓度>50 ng/μL的DNA样品检测结果见表 1。24例样本中16例芯片检测成功,8例失败。芯片成功样本与失败样本的D260/D280值分别为1.860±0.063与1.841±0.825,差异无统计学意义(P=0.444);而两组的D260/D230值分别为2.37(0.91~4.95)、0.23(0.15~0.45),差异有统计学意义(Z=-3.920,P < 0.001)。
|
|
表 1 24例DNA样本浓度及芯片检测结果 Tab 1 Concentrations and microarray detection results of 24 DNA samples |
2.3 PCR及基因分型结果
第二阶段1 122例NanoDrop及PicoGreen检测浓度均大于50 ng/μL的DNA样品中979例PCR成功,143例失败。对979例PCR成功的DNA样本行基因分型检测,结果显示其Call rate值达标率为100%。将1 122例样品按照PCR结果分为两组,非参数检验结果显示,PCR成功与失败两组DNA样本的D260/D230值分别为1.45(0.09~18.61)和0.54(0.08~15.17),差异有统计学意义(Z=-5.983, P < 0.01)。
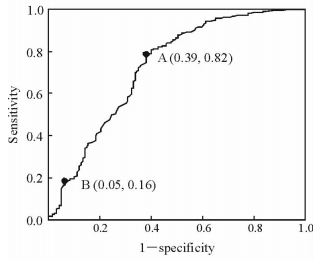
2.4 D260/D230值对PCR结果的鉴别能力通过绘制ROC曲线评估D260/D230对PCR结果的鉴别能力,结果(图 1)显示,ROC曲线的AUC为0.727;其中最佳诊断点(敏感度和特异度最大)的D260/D230值为89%(敏感度为82%,特异度为61%)。ROC曲线中特异度为95%(敏感度16%)时对应的D260/D230值为2.305。
|
图 1 D260/D230预测PCR结果的ROC曲线 Fig 1 ROC curve of PCR results predicted by D260/D230 Point A: Best diagnostic point; Point B: Point of specificity 95%. PCR: Polymerase chain reaction; ROC: Receiver operating characteristic |
3 讨论
GWAS及二代测序实验的影响因素有多种,如诊断明确的病例和对照组、多项检测的控制、人群分层的控制[8]、有明确表型的研究人群的特点[9]以及DNA样品的数量和质量等[10]。以GWAS为例,由于芯片非常昂贵,所以在进行芯片检测之前确保DNA样本的质量非常重要。
目前遗传学研究大多采用NanoDrop和PicoGreen检测DNA样品的质量。NanoDrop检测方法简单、经济。DNA在波长260 nm左右有最大吸收峰,而蛋白质的吸收峰在280 nm左右[5],因此可通过D260/D280值及D260/D230值检测DNA浓度;但RNA、单链DNA (ssDNA)和双链DNA (dsDNA)均在260 nm处有最大吸收峰,导致整体的D260值可能偏高,影响检测结果。PicoGreen是一种选择性结合dsDNA的荧光染料,与dsDNA结合时其荧光急剧增强,而未结合染料几乎不发出荧光,因此被用来检测DNA浓度;且PicoGreen稳定不易脱色,允许暴露较长的时间[6]。PCR-扩增管家基因(GAPDH)的方法常被用于确定样本基因组DNA的完整性及纯度[8, 11],因此本研究利用PCR-扩增管家基因以提高DNA芯片检测的成功率。
NanoDrop和PicoGreen测定DNA浓度时,纯度高的核酸其D260/D280值一般为1.8~2.0。但该比值受pH和离子浓度影响,酸性溶液一般会使其降低0.2~0.3,而碱性溶液则可能使其上升0.2~0.3;异常的D260/D280值表示样品中可能含有蛋白质、苯或其他在280 nm附近有吸收的污染物。D260/D230值是另一用于检测DNA纯度的指标,纯度高的核酸其D260/D230值一般为1.8~2.2,此值明显偏小时表明样品可能被碳水化合物(糖类)、盐类或有机溶剂污染,需要纯化样品或进一步优化核酸提取方法[12]。大多数研究中,一般使用D260/D280值评价DNA的纯度,而D260/D230值则被认为是次要的[13]。但近年来,涉及到DNA质控的遗传学研究越来越强调D260/D230值的价值。Solomon等[14]提取海水微生物DNA时发现,采用新的提取方法后,样本的D260/D230值可改善2.3%~45%,而D260/D280值未见明显改变,抽提样本可降低22.5%~34.5%的蛋白质污染,DNA合格率及16S RNA的PCR成功率均显著提高。本研究第一阶段中PCR结果阳性与阴性两组的D260/D280值差异无统计学意义,而D260/D230值差异有统计学意义(P < 0.001)。造成这种差异的原因可能是抽提的部分DNA样本中含有较多的碳水化合物(糖类)、盐类等污染物,这些污染物对230 nm的波段吸收较好而对280 nm波段吸收不佳,即表现为D260/D280值无明显变化而D260/D230值偏低。表明在衡量样本纯度时D260/D230值可能有重要意义,在检测样本时忽视该值也可能无法确保样本的质量。
本研究中,ROC曲线的AUC为0.727,表明所建立的D260/D230值对PCR结果模型有较好的预测效能。本研究得到的最佳诊断点D260/D230值为0.89,低于正常的D260/D230值范围(1.8~2.2),推测原因可能为当DNA混有一定程度的杂质时PCR仍可成功。但芯片对DNA纯度要求较高,故在芯片基因分型前的质检阶段,特异度(即D260/D230值预测PCR成功的可靠性)较敏感度更为重要,它可以避免因样本质量问题而导致的芯片浪费。因此本研究选择并推荐特异度为95%的诊断点(D260/D230值为2.305)为衡量标准,D260/D230值≥2.305且浓度合格的样本,其PCR-扩增管家基因成功的可能性超过95%,样本纯度较高,基本满足芯片检测要求。
在本研究第一阶段,即使样本经过NanoDrop及PicoGreen检测显示浓度已达到50 ng/μL,但仍有部分芯片检测失败。而第二阶段中经过NanoDrop、PicoGreen及PCR质检的样本芯片检测成功率为100%,表明与单用NanoDrop或PicoGreen的质检方法相比,其联合应用PCR检测能更大程度地确保GWAS的成功。
综上所述,在GWAS的DNA质检过程中,尽管NanoDrop检测中D260/D230值的正常范围为1.8~2.2,但当D260/D230 < 2.305时,DNA样本仍可能有污染,有可能导致后续遗传学研究失败,此时应进行PCR-扩增管家基因质检以确保GWAS芯片检测成功;当D260/D230≥2.305时则可认为PCR成功率超过95%,样本纯度基本满足芯片检测要求,故不需再进行PCR质检,可提高样本质检效率。本研究不足之处在于样本量有限,且PCR成功组与失败组的数量差异较大,仍需更大的多中心数据予以支持。
| [1] | JIANG L, YIN J, YE L, YANG J, HEMANI G, LIU A J, et al. Novel risk loci for rheumatoid arthritis in Han Chinese and congruence with risk variants in Europeans[J]. Arthritis Rheumatol, 2014, 66: 1121–1132. DOI: 10.1002/art.v66.5 |
| [2] | OKADA Y, WU D, TRYNKA G, RAJ T, TERAO C, IKARI K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery[J]. Nature, 2014, 506: 376–381. |
| [3] | DAVIDSON S I, JIANG L, CORTES A, WU X, GLAZOV E A, DONSKOI M, et al. Brief report:high-throughput sequencing of IL23R reveals a low-frequency, nonsynonymous single-nucleotide polymorphism that is associated with ankylosing spondylitis in a Han Chinese population[J]. Arthritis Rheum, 2013, 65: 1747–1752. DOI: 10.1002/art.37976 |
| [4] | LAURIE C C, DOHENY K F, MIREL D B, PUGH E W, BIERUT L J, BHANGALE T, et al. Quality control and quality assurance in genotypic data for genome-wide association studies[J]. Genet Epidemiol, 2010, 34: 591–602. DOI: 10.1002/gepi.20516 |
| [5] | LEE J H, PARK Y, CHOI J R, LEE E K, KIM H S. Comparisons of three automated systems for genomic DNA extraction in a clinical diagnostic laboratory[J]. Yonsei Med J, 2010, 51: 104–110. DOI: 10.3349/ymj.2010.51.1.104 |
| [6] | AHN S J, COSTA J, EMANUEL J R. PicoGreen quantitation of DNA:effective evaluation of samples pre-or post-PCR[J]. Nucleic Acids Res, 1996, 24: 2623–2625. DOI: 10.1093/nar/24.13.2623 |
| [7] | VAN DER LINDEN S, VALKENBURG H A, CATS A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria[J]. Arthritis Rheum, 1984, 27: 361–368. DOI: 10.1002/(ISSN)1529-0131 |
| [8] | PEARSON T A, MANOLIO T A. How to interpret a genome-wide association study[J]. JAMA, 2008, 299: 1335–1344. DOI: 10.1001/jama.299.11.1335 |
| [9] | MARIAN A J. Molecular genetic studies of complex phenotypes[J]. Transl Res, 2012, 159: 64–79. DOI: 10.1016/j.trsl.2011.08.001 |
| [10] | STEINBERG K, BECK J, NICKERSON D, GARCIA-CLOSAS M, GALLAGHER M, CAGGANA M, et al. DNA banking for epidemiologic studies:a review of current practices[J]. Epidemiology, 2002, 13: 246–254. DOI: 10.1097/00001648-200205000-00003 |
| [11] | USMAN T, YU Y, LIU C, FAN Z, WANG Y. Comparison of methods for high quantity and quality genomic DNA extraction from raw cow milk[J]. Genetics Mol Res, 2014, 13: 3319–3328. DOI: 10.4238/2014.April.29.10 |
| [12] | HANSEN H M, WIEMELS J L, WRENSCH M, WIENCKE J K. DNA quantification of whole genome amplified samples for genotyping on a multiplexed bead array platform[J]. Cancer Epidemiol Biomarkers Prev, 2007, 16: 1686–1690. DOI: 10.1158/1055-9965.EPI-06-1024 |
| [13] | DESJARDINS P, CONKLIN D. NanoDrop microvolume quantitation of nucleic acids[J/OL]. J Vis Exp, 2010:e2565. doi:10.3791/2565. |
| [14] | SOLOMON S, KACHIPRATH B, JAYANATH G, SAJEEVAN T P, BRIGHT SINGH I S, PHILIP R. High-quality metagenomic DNA from marine sediment samples for genomic studies through a preprocessing approach[J/OL]. 3 Biotech, 2016, 6:160. doi:10.1007/s1205-016-0482-y. |
 2017, Vol. 38
2017, Vol. 38


