非编码RNA(non-coding RNA,ncRNA)在细胞各项生物过程中发挥着不可忽视的作用,被喻为生命的“暗物质”。近年随着RNA测序技术和生物信息学的发展,研究者发现了非编码RNA家族的另一重要成员——环状RNA(circular RNA,circRNA),同时其产生机制、生物学特性和功能不断更新,相关临床研究相继开展,现已成为疾病研究的热点之一[1]。环状RNA的发现已近40年,早在1976年,人们发现类病毒由环形的RNA分子构成[2];1979年,Hsu和Coca-Prados[3]利用电镜在HeLa细胞中发现环状RNA占总RNA含量的1%~2%;1991年,Nigro等[4]在人体细胞中发现了DCC促癌基因存在外显子倒向连接现象,后证明这种连接方式的产物是环状RNA[5]。目前,环状RNA已由“剪切副产物”成为非编码RNA研究领域的新星。本文对真核细胞内源性环状RNA的研究进展及其与疾病的关联作一综述。
1 环状RNA的生成与特性真核细胞内源性环状RNA是一类环形的基因转录产物,无游离的5′端或3′端,核酸分子间形成闭合3′-5′磷酸二酯键(内含子套索RNA成环处碱基以2′-5′磷酸二酯键结合),构成单链环形结构。环状RNA通常由1~5个外显子或1~2个内含子组成,同时也可含有基因间区或非编码区成分[6, 7, 8]。仅由内含子构成的环状RNA称为内含子环状RNA(circular intron RNA,ciRNA),同时具有外显子与内含子序列的环状RNA称为外显子-内含子环状RNA(exon-intron circRNAs,EIciRNAs)[9]。在计算机辅助下,将转录本测序结果与已知基因组序列相匹配,鉴别出反向剪切位点,便可预测环状RNA的存在[10]。
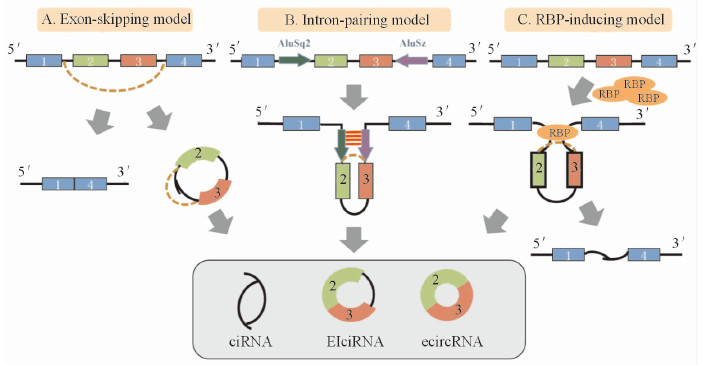
1.1 环状RNA的产生及调控机制环状RNA的产生机制和过程尚未彻底明确。现有研究显示,环状RNA由前体mRNA(pre-mRNA)反向剪切而成(图 1A)[8, 11, 12]。在Jeck等[8]提出的RNA成环模型中,剪切供体(splice donor)和剪切受体(splice acceptor)分别以外显子跳读(exon-skipping)和内含子互补的方式缩小空间距离,反向剪切形成环状RNA。环状RNA形成受外显子侧翼序列(图 1B)或RNA结合蛋白(RNA binding protein,RBP,图 1C)的调控[9, 11, 13, 14, 15, 16]。环状RNA侧翼内含子长度明显高于全基因组平均值[17],其Alu元件(Alu element)数量成倍增加,更易于形成互补结构。例如ZKSCAN1基因第2外显子上游的AluSq2元件中,有87%的碱基能与第3外显子下游的AluSz元件互补,提高第2、3外显子成环效率[18]。RNA结合蛋白Quaking属于STAR家族,能特异识别NACUAAY核心(core) 及UAAY半位点(half-site)[19],牵拉结合点相互缩小空间距离,促进成环。SMARCA5基因第15、16外显子所形成的环状RNA(编号hsa_circ_0001445)侧翼序列中存在丰富的UAAY半位点,将此类位点突变后,环状RNA生成显著减少。另外,RNA剪辑因子(RNA-editing factor)ADAR1(adenosine deaminase 1)能对环状RNA的形成起到负性调节作用。ADAR1可阻止外显子侧翼序列互补结构的形成,从而抑制外显子环化。实验证实,敲除HEK293细胞及人类成神经细胞瘤细胞SH-SY5Y中ADAR基因后,多种环状RNA表达量均有不同程度的提高[20]。环状RNA还受到miRNA调控,miR-671可通过碱基互补的方式介导AGO2蛋白对ciRS-7(circular RNA sponge for miR-7;由CDR1反义链产生的环状RNA,可吸附miR-7)分子进行剪切[21]。
|
图 1 环状RNA形成模型 Fig 1 Models of circRNA biogenesis vessels A: Exon-skipping of pre-mRNA produces linear mRNAs and forms circles of the rest; B: Introns with complementary ALU repeats form circles through base-pairing; C: RNA binding proteins binding to the target sites in the flanking introns induce circularization.circRNA: Circular RNA; ciRNA: Circular intron RNA; EIciRNA: Exon-intron circRNA; ecircRNA: Circular exon RNA |
随着研究的不断深入,环状RNA的诸多特性引起了人们的注意,其中受到公认及较为重要的包括以下4点:
(1)环状RNA种类繁多,广泛、大量存在于真核细胞。迄今,全球的学者已经在包括人、鼠、斑马鱼、果蝇、线虫、酵母菌、水稻在内的动植物及真菌细胞内预测出超过10万种环状RNA[6, 8, 20, 22, 23, 24, 25]。对小鼠的研究发现,有20%的蛋白质编码基因能够产生环状RNA[26],并且部分基因可产生多种环状RNA亚型。Salzman等[23]在真核细胞内检测到环状RNA的含量(分子数量)占polyA阳性RNA分子的1%,且在其线性异构体中占5%~10%的比例。 Rims2基因产生的环状RNA含量甚至达到其线性mRNA表达量的20倍[20],这有力反驳了环状RNA是剪切副产物的假说,标示着环状RNA可能在真核细胞RNA分子中占有不可忽视的重要地位。
(2)环状RNA生物学性质稳定。环状RNA分子的稳定性很大程度上来源于其环形结构导致的核酸酶逃逸。RNA酶(RNase)R是一种高效核酸外切酶,能识别RNA分子游离3′端,并向5′端水解靶RNA,可有效水解单链线性RNA分子[27]。由于环状RNA首尾相接,缺乏游离的3′端,故不为RNase R识别并水解。此外,RNase Ⅱ、多核苷酸磷酸化酶、寡核糖核酸酶等常见核酸酶均不能有效水解环状RNA[28]。细胞内阻断环状RNA的生成,其含量在一定时间内无明显下降,是环状RNA稳定性较高的另一证据[8]。研究人员在人体唾液上清中检测到超过400种环状RNA[29],这些环状RNA的亲本基因与炎症反应、细胞骨架形成、细胞运动、T细胞极性形成以及整联蛋白介导的信号通路等相关。此外,Li等[30]在人体血清外泌体中发现了超过1 000种环状RNA。这些证据预示着环状RNA可能作为良好的信息载体,在细胞内及细胞间具有信息传递功能。
(3)环状RNA具有时空特异性。例如,ciRS-7在脑组织中表达较高,而在非神经组织中表达量则较低[31];DOCK1基因表达的环状RNA在乳腺癌细胞系MCF-7中呈高表达,而在肺癌细胞系A549中则几乎不表达[23]。在不同发育时期的线虫胚胎细胞中,环状RNA种类及相对表达量均存在显著差异[26, 31]。另外,通过荧光原位杂交技术可观察到环状RNA的分布具有明显细胞质或细胞核聚集性,例如ciRS-7主要存在于细胞质,而circEIF3J和ci-ankrd52则仅聚集于细胞核[9, 31, 32]。时空特异性是环状RNA具有特定功能的证据,也是将其运用于开发临床疾病诊断标记物的基础。
(4)环状RNA具有生物进化保守性。Jeck等[8]在人体成纤维细胞中发现2 121种环状RNA能与小鼠基因组匹配,其中457个被匹配的小鼠基因能产生相同的环状RNA。Guo等[33]发现能产生环状RNA的小鼠基因所对应的人类同源基因有66%的可能产生环状RNA;反之,对于不能产生环状RNA的小鼠基因,其人类同源基因产生环状RNA的比例只有19%。尽管如此,环状RNA的保守性到底是基因保守的必然结果,还是因具重要功能而受到精确调控形成,尚需进一步研究证明。
2 环状RNA与其他分子的相互作用及功能 2.1 环状RNA与miRNA的相互作用环状RNA参与了复杂的RNA-RNA相互作用网络,在基因转录后调控中发挥作用。部分环状RNA包含若干miRNA结合位点,具有miRNA海绵功能(miRNA sponge)。来源于小鼠Y染色体性别决定区的环状RNA分子circSry,是第1个被发现能与RNA相互作用的环状RNA,序列分析推测该环状RNA能够与16个miR-138分子结合,抑制miR-138分子对其下游靶标的作用[34]。另外,作为环状RNA的代表分子之一,小脑退行性相关蛋白基因(CDR1)反义链转录的环状RNA分子CDR1as/ciRS-7可高效吸附miR-7,抑制其对目标基因mRNA的沉默[31, 35, 36],同时也受miR-671介导的RNA降解。通过生物信息学分析已发现上万种环状RNA具有miRNA吸附功能[37],但仅有少数得到了验证,因此,笼统地将环状RNA的功能概括为RNA海绵仍缺乏充足的证据。
2.2 环状RNA与蛋白质的相互作用环状RNA可直接与蛋白质结合,也可通过RNA介导间接与蛋白质发生关联,影响蛋白质功能。例如,RNA剪切因子MBL可结合其亲本基因第2外显子,促进其环化形成circMbl;同时,circMbl能与MBL结合,降低MBL有效浓度,减少circMbl生成[38]。内含子-外显子环状RNA circEIF3J与U1 RNA结合,促进U1小核核糖核蛋白(U1 snRNP)复合体与RNA聚合酶Ⅱ结合,增强亲本基因转录[9]。circ-Foxo3与激酶抑制蛋白(p21)及细胞分裂蛋白激酶2(CDK2)形成复合体,抑制CDK2对细胞分裂的促进作用,阻碍细胞周期进展[39]。
2.3 环状RNA的翻译功能水稻黄斑驳病毒scRYMV[40]以及人工合成的具有内部核糖体进入位点(internal ribosome entry site,IRES)的环形RNA分子在细胞内能够翻译成蛋白质[41, 42, 43]。因此,环形结构并非影响RNA编码的决定性因素。但是,目前在真核细胞内尚无内源性环状RNA可直接翻译出蛋白质的证据:(1)未检测到环状RNA与核糖体结合[8];(2)在A549细胞系内,蛋白质谱分析未发现与环状RNA序列对应的蛋白质[44]。
3 环状RNA相关临床研究进展环状RNA与疾病的发生和发展有密切关联[36, 45],是一种充满前景的生物标记物甚至治疗靶点[46],成为继miRNA及长链非编码RNA(LncRNA)之后又一临床疾病研究热点。有学者预测ciRS-7与miR-7的相互作用可能影响到miR-7靶基因(EGFR、PIK3CD、mTOR等)的表达,从而与恶性胶质瘤、胃癌、肝癌等疾病的发生、发展相关联[45, 46, 47, 48, 49]。Li等[50]发现食管癌组织中环状ITCH(cir-ITCH)的表达量显著低于正常组织,在人工构建的荷瘤小鼠模型中,外源过表达cir-ITCH可抑制食管癌肿瘤细胞生长,证明环状RNA与肿瘤的发生、发展具有直接关联,为更多将环状RNA投入临床研究的学者树立了信心。Bachmayr-Heyda等[51]通过对直肠癌组织转录组测序进行分析,发现环状RNA表达量在肿瘤组织及肿瘤细胞系中具有整体下降的趋势,并且特定环状RNA与其线性异构体的比值在结直肠癌与癌旁组织中具有显著差异,这为探索肿瘤发病机制提供了新的思路。
除肿瘤外,环状RNA与衰老、炎症等生理、病理现象的相关研究也不断兴起[52, 53]。对环状RNA在疾病中差异性表达的测定及其机制的研究或许能够帮助临床研究者诊断疾病、评估疗效、分析病机及开发新药物治疗靶点。Du等[52]发现,circ-Foxo3在老年患者及小鼠的心肌细胞内表达较高,干扰circ-Foxo3的表达可以抑制小鼠胚胎成纤维细胞的衰老。随后,更深入的研究证明circ-Foxo3与抗衰老相关蛋白ID-1和E2F1及抗应激蛋白FAK和HIF1a结合,抑制蛋白抗衰老功能,导致心肌衰老。此外,目前已有多项研究分别证明了如骨关节炎[53]、大脑缺血再灌注损伤[54]、动脉粥样硬化[55]、子痫前期[56]等非肿瘤性疾病的临床组织、血液样本或疾病细胞模型中存在环状RNA的异常表达。环状RNA凭借其特殊的生物学特性及功能,与肿瘤、生物标记物、衰老等研究热点相结合[17, 30, 52],参与到生物与医学研究的各个环节,成为基础科研与临床研究相互转化的桥梁。
4 展望环状RNA研究手段日趋成熟,目前Circbase、Circ2Traits、CircNet等数据库已收录了近10万种环状RNA测序结果,并能预测circRNA-miRNA-mRNA相互作用,帮助研究者系统地研究环状RNA[57, 58, 59]。同时,人工构建环状RNA或干扰环状RNA的方法不断涌现并趋于成熟,使人为调控细胞内环状RNA的表达成为可能,有利于进一步探索环状RNA的功能[15, 31]。分析环状RNA相关文献的发表情况可知,环状RNA与疾病的相关研究正由“提出假设”向“实践证明”转变,将环状RNA运用于临床诊疗工作或成为转化医学与精准医疗的新落脚点。相信随着研究技术的发展与普及,环状RNA将带给医学和科学界更多的惊喜与突破。
| [1] | 夏世金, 高 文, 胡明冬, 刘露梅, 邰先桃. 环状RNA的研究现状及展望[J/CD].中华肺部疾病杂志(电子版), 2014, 7:670-673. |
| [2] | Sanger H L, Klotz G, Riesner D, Gross H J, Kleinschmidt A K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures[J]. Proc Natl Acad Sci USA, 1976, 73: 3852-3856. |
| [3] | Hsu M T, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells[J]. Nature, 1979, 280: 339-340. |
| [4] | Nigro J M, Cho K R, Fearon E R, Kern S E, Ruppert J M, Oliner J D, et al. Scrambled exons[J]. Cell, 1991, 64: 607-613. |
| [5] | Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules[J]. FASEB J, 1993, 7: 155-160. |
| [6] | Salzman J, Gawad C, Wang P L, Lacayo N, Brown P O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types[J]. PLoS One, 2012, 7: e30733. |
| [7] | Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea[J]. Nucleic Acids Res, 2012, 40: 3131-3142. |
| [8] | Jeck W R, Sorrentino J A, Wang K, Slevin M K, Burd C E, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats[J]. RNA, 2013, 19: 141-157. |
| [9] | Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus[J]. Nat Struct Mol Biol, 2015, 22: 256-264. |
| [10] | Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification[J]. Genome Biol, 2015, 16: 4. |
| [11] | Barrett S P, Wang P L, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor[J]. Elife, 2015, 4: e07540. |
| [12] | Ebbesen K K, Kjems J, Hansen T B. Circular RNAs: identification, biogenesis and function[J]. Biochim Biophys Acta, 2016, 1859: 163-168. |
| [13] | Vicens Q, Westhof E. Biogenesis of circular RNAs[J]. Cell, 2014, 159: 13-14. |
| [14] | Ivanov A, Memczak S, Wyler E, Torti F, Porath H T, Orejuela M R, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals[J]. Cell Rep, 2015, 10: 170-177. |
| [15] | Conn S J, Pillman K A, Toubia J, Conn V M, Salmanidis M, Phillips C A, et al. The RNA binding protein quaking regulates formation of circRNAs[J]. Cell, 2015, 160: 1125-1134. |
| [16] | Zhang X O, Wang H B, Zhang Y, Lu X, Chen L L, Yang L. Complementary sequence-mediated exon circularization[J]. Cell, 2014, 159: 134-147. |
| [17] | Westholm J O, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation[J]. Cell Rep, 2014, 9: 1966-1980. |
| [18] | Liang D, Wilusz J E. Short intronic repeat sequences facilitate circular RNA production[J]. Genes Dev, 2014, 28: 2233-2247. |
| [19] | Galarneau A, Richard S. Target RNA motif and target mRNAs of the quaking STAR protein[J]. Nat Struct Mol Biol, 2005, 12: 691-698. |
| [20] | Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed[J]. Mol Cell, 2015, 58: 870-885. |
| [21] | Hansen T B, Wiklund E D, Bramsen J B, Villadsen S B, Statham A L, Clark S J, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA[J]. EMBO J, 2011, 30: 4414-4422. |
| [22] | Chen I, Chen C Y, Chuang T J. Biogenesis, identification, and function of exonic circular RNAs[J]. Wiley Interdiscip Rev RNA, 2015, 6: 563-579. |
| [23] | Salzman J, Chen R E, Olsen M N, Wang P L, Brown P O. Cell-type specific features of circular RNA expression[J]. PLoS Genet, 2013, 9: e1003777. |
| [24] | Wang P L, Bao Y, Yee M C, Barrett S P, Hogan G J, Olsen M N, et al. Circular RNA is expressed across the eukaryotic tree of life[J]. PLoS One, 2014, 9: e90859. |
| [25] | Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, et al. Transcriptome-wide investigation of circular RNAs in rice[J]. RNA, 2015, 21: 2076-2087. |
| [26] | You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity[J]. Nat Neurosci, 2015, 18: 603-610. |
| [27] | Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs[J]. Int J Mol Sci, 2014, 15: 9331-9342. |
| [28] | Suzuki H, Zuo Y, Wang J, Zhang M Q, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing[J]. Nucleic Acids Res, 2006, 34: e63. |
| [29] | Bahn J H, Zhang Q, Li F, Chan T M, Lin X, Kim Y, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva[J]. Clin Chem, 2015, 61: 221-230. |
| [30] | Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis[J]. Cell Res, 2015, 25: 981-984. |
| [31] | Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency[J]. Nature, 2013, 495: 333-338. |
| [32] | Zhang Y, Zhang X O, Chen T, Xiang J F, Yin Q F, Xing Y H, et al. Circular intronic long noncoding RNAs[J]. Mol Cell, 2013, 51: 792-806. |
| [33] | Guo J U, Agarwal V, Guo H, Bartel D P. Expanded identification and characterization of mammalian circular RNAs[J]. Genome Biol, 2014, 15: 409. |
| [34] | Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis[J]. Cell, 1993, 73: 1019-1030. |
| [35] | Hansen T B, Jensen T I, Clausen B H, Bramsen J B, Finsen B, Damgaard C K, et al. Natural RNA circles function as efficient microRNA sponges[J]. Nature, 2013, 495: 384-388. |
| [36] | Hansen T B, Kjems J, Damgaard C K. Circular RNA and miR-7 in cancer[J]. Cancer Res, 2013, 73: 5609-5612. |
| [37] | Thomas L F, Sætrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites[J]. Bioinformatics, 2014, 30: 2243-2246. |
| [38] | Ashwal-Fluss R, Meyer M, Pamudurti N R, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing[J]. Mol Cell, 2014, 56: 55-66. |
| [39] | Du W W, Yang W, Liu E, Yang Z, Dhaliwal P, Yang B B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2[J]. Nucleic Acids Res, 2016 Feb 9. pii: gkw027. [Epub ahead of print] |
| [40] | Abouhaidar M G, Venkataraman S, Golshani A, Liu B, Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt[J]. Proc Natl Acad Sci USA, 2014, 111: 14542-14547. |
| [41] | Chen C Y, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs[J]. Science, 1995, 268: 415-417. |
| [42] | Perriman R, Ares M Jr. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo[J]. RNA, 1998, 4: 1047-1054. |
| [43] | Conn S J, Pillman K A, Toubia J, Conn V M, Salmanidis M, Phillips C A, et al. The RNA binding protein quaking regulates formation of circRNAs[J]. Cell, 2015, 160: 1125-1134. |
| [44] | Salzman J, Chen R E, Olsen M N, Wang P L, Brown P O. Cell-type specific features of circular RNA expression[J]. PLoS Genet, 2013, 9: e1003777. |
| [45] | Peng L, Yuan X Q, Li G C. The emerging landscape of circular RNA ciRS-7 in cancer (review)[J]. Oncol Rep, 2015, 33: 2669-2674. |
| [46] | Li J, Yang J, Zhou P, Le Y, Zhou C, Wang S, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications[J]. Am J Cancer Res, 2015, 5: 472-480. |
| [47] | Li P, Chen S, Chen H, Mo X, Li T, Shao Y, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer[J]. Clin Chim Acta, 2015, 444: 132-136. |
| [48] | Song X, Zhang N, Han P, Moon B S, Lai R K, Wang K, et al. Circular RNA profile in gliomas revealed by identification tool UROBORUS[J]. Nucleic Acids Res, 2016 Feb 11. pii: gkw075. [Epub ahead of print] |
| [49] | Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma[J]. Cancer Biomark, 2016, 16: 161-169. |
| [50] | Li F, Zhang L, Li W, Deng J, Zheng J, An M, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway[J]. Oncotarget, 2015, 6: 6001-6013. |
| [51] | Bachmayr-Heyda A, Reiner A T, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, et al. Correlation of circular RNA abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues[J]. Sci Rep, 2015, 5: 8057. |
| [52] | Du W W, Yang W, Chen Y, Wu Z K, Foster F S, Yang Z, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses[J]. Eur Heart J, 2016 Feb 11. pii: ehw001. [Epub ahead of print] |
| [53] | Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 'Sponge’ in human cartilage degradation[J]. Sci Rep, 2016, 6: 22572. |
| [54] | Lin S, Ye S, Long Y, Fan Y, Mao H F, Chen M T, et al. Circular RNA expression alterations are involved in OGD/R-induced neuron injury[J]. Biochem Biophys Res Commun, 2016, 471: 52-56. |
| [55] | Burd C E, Jeck W R, Liu Y, Sanoff H K, Wang Z, Sharpless N E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk[J]. PLoS Genet, 2010, 6: e1001233. |
| [56] | Zhang Y, Yang H, Long Y, Li W L. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia[J]. BJOG, 2016 Feb 5. doi: 10.1111/1471-0528.13897. [Epub ahead of print] |
| [57] | Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits[J]. Front Genet, 2013, 4: 283. |
| [58] | Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs[J]. RNA, 2014, 20: 1666-1670. |
| [59] | Liu Y C, Li J R, Sun C H, Andrews E, Chao R F, Lin F M, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data[J]. Nucleic Acids Res, 2015, 44(D1): D209-D215. |
 2016, Vol. 37
2016, Vol. 37


