2. University of Chinese Academy of Science, Beijing 100049, China
2. 中国科学院大学, 北京 100049
The two-component system (TCS) plays an important role in bacteria signal transduction. It is mainly found in prokaryotes, and in a small number of eukaryotes, but it is rarely found in animals and plants and notably absent in human body[1]. TCSs are involved in various signal transduction pathways such as environmental Pi concentration, pH, osmotic pressure, temperature, and so on[2]. The typical TCS comprises a sensor histidine kinase (HK) and a response regulator (RR)[3]. The signal transduction pathway is mediated by the phosphorylation and dephosphorylation between these proteins[4, 5]. The Mg2+ is an essential component of the phosphorylation of numerous response regulators through coordination with the phosphoryl group and protein sidechains to stabilize the active conformation of RR, that is extremely conserved in many TCSs[6, 7].
Nuclear magnetic resonance (NMR) spectroscopy is powerful to explore the conformational dynamics of RRs in their apo state without Mg2+ and phosphoryl group. The 1H-15N HSQC NMR spectrum provides a residue-specific fingerprint of the overall protein conformation. By monitoring changes in chemical shift and/or line broadening of previously assigned resonances in the protein spectrum, it would provide abundant information of conformation exchanges[8]. Based on relaxation experiments, it has been characterized that NtrC has an exchange between inactive and active conformations, and both states are populated in unphosphorylated NtrC[9]. However, the minor active conformation is hard to be visualized by traditional NMR experiments directly.
In order to visualize the minor active conformation preexisting in the apo state of RR directly, we use PhoB as a model. PhoB is a response regulator of the PhoR/PhoB signal transduction system from Escherichia coli and controls the expression of over 40 genes related to phosphate assimilation[10, 11]. It consists of a N-terminal receiver domain (PhoBN) that contains the conserved phosphorylation site and a C-terminal effector domain that performs the output function[12-14]. The crystal structure of active and inactive PhoBN have been reported, respectively[13].
In previous studies, we have found that apo PhoBN has monomer-to-dimer equilibrium[13] and the dimer has multiple conformational changes in solution. The NMR spectrum of PhoBN gives inhomogeneous signals and makes it hard to investigate. It has been reported that F20 stabilizes one of the dimer interfaces (α1-α5) through hydrophobic interactions with V21 and the aromatic rings of the Q24 side chain[15]. We replaced F20 with an aspartate residue (PhoBNF20D) which disrupted the α1-α5 dimer interface, and demonstrated that PhoBNF20D becomes a monomer in the solution and has the same phosphorylation property as the wild type PhoBN[16]. The relaxation parameters of the backbone amide nitrogens of apo PhoBNF20D were measured and the overall rotation correlation time (τc) was calculated. It gives the value of 12.87 ns which corresponds to the molecular weight of the monomer. Therefore, the mutant PhoBNF20D can be used to explore the conformational exchange between active and inactive state simply at monomer state. Moreover, beryllium fluoride (BeF3-) is widely used as a phosphoryl analogue in macromolecular studies and it can be applied to stabilize the active conformation, meanwhile both bound and unbound BeF3- can be detected by 19F NMR directly[17].
Here we analyze the interaction between PhoBNF20D and BeF3- with or without Mg2+ using NMR. We find that apo PhoBNF20D has a pre-active conformation in solution and BeF3- can stabilize this conformation without Mg2+. The minor pre-active conformation was visualized by 1H-15N HSQC and 19F NMR spectra directly. Meanwhile, Mg2+ can shift the equilibrium between inactive and active conformation and makes PhoBNF20D completely become active conformation. The Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion experiments verify that there is an exchange between the inactive and active conformations of apo PhoBNF20D. Our results provide direct evidence of the existing of minor active conformation in the apo state of RR, and a better understanding of conformational exchange during the phosphorylation process of response regulators.
1 Experimental method 1.1 Expression of PhoBNF20DThe E.coli phoB gene was cloned into the pET-28a vector (Novagen) using the NdeI and XhoI restriction enzyme sites to express PhoBN (1~125 residue) protein with thrombin-cleavable His6 tag[18] at the N-terminus. The PhoBNF20D mutant was produced by polymerase chain reaction (PCR). The constructed plasmid was transformed into E.coli BL21(DE3) strain for protein expression and purification. The expression cell stock was grown in LB medium containing 50 μg/mL kanamycin at 37 ℃ overnight. 15N-labeled PhoBNF20D was expressed in 500 mL M9 media supplemented with 0.5 g 15NH4Cl (Cambridge Isotope Laboratories, Andover, MA, USA). When OD600 of the culture reached to 0.7~0.8, the cells were induced by 1 mmol/L isopropyl thio β-D-1-galactopyranoside at 20 ℃ for 20~21 h before harvested.
1.2 Protein purification and sample preparationThe cultured cells were resuspended in lysate buffer (containing 50 mmol/L Tris, 300 mmol/L NaCl, and 20 mmol/L imidazole), and disrupted by sonication on ice. The cell debris was removed by centrifugation (48 384 g, 40 min, 4 ℃). Liquid supernatant was filtrated and then loaded onto the Ni2+-NTA column which was equilibrated with buffer A (containing 50 mmol/L Tris, 300 mmol/L NaCl, and 20 mmol/L imidazole). PhoBNF20D was eluted by 70% (v/v) buffer B (containing 50 mmol/L Tris, 300 mmol/L NaCl, and 500 mmol/L imidazole). The His6-fused protein was cleaved by bovine thrombin (2~3 h, room temperature). The redundant bovine thrombin was removed by benzamidine column. After that, the PhoBNF20D protein was loaded onto the Ni2+-NTA column again to remove the uncleaved protein and then further purified by Superdex 75 (buffer containing 50 mmol/L Tris, 100 mmol/L NaCl, and 5 mmol/L dithiothreitol) and desalting column.
1.3 NMR spectroscopyAll the NMR samples were prepared in the buffer containing 20 mmol/L HEPES (pH 7.0), 100 mmol/L NaCl, 5 mmol/L DTT and 10% (v/v) D2O and 90% (v/v) H2O. All protein NMR experiments were performed on 0.3 mmol/L or 0.8 mmol/L 15N-labeled PhoBNF20D. In addition, samples for 19F NMR and 1H-15N HSQC titration experiments consisted of 20 mmol/L HEPES, 100 mmol/L NaCl, 5 mmol/L DTT, 1.5 mmol/L BeCl2 and 15 mmol/L NaF, with or without 10 mmol/L Mg2+, pH 7.0.
1H-15N HSQC experiments were performed on Bruker 600 MHz equipped with 1H/19F(13C)/15N channels and triple-resonance cryo-probes with pulsed-field gradients at 298 K. 1H-15N HSQC spectra were acquired with 1H carrier set coincident with the water resonance and 15N frequency set to δN 119; spectra widths of F2 (1H) and F1 (15N) dimensions were 7 183.908 Hz and 2 188.284 Hz, respectively. The bandwidth of 1D 19F spectrum was 40 760.871 Hz and the chemical shifts were calibrated according to our previous study[17]. Mg2+ titration experiments in the presence of BeF3- were carried out with the concentrations of Mg2+ at 0.05, 0.10, 0.30 and 0.50 mmol/L, respectively.
A set of constant-time 15N CPMG relaxation dispersion experiments was performed on Bruker 850 MHz NMR spectrometer equipped with 1H/19F(13C)/15N channels and triple-resonance cryo-probes with pulsed-field gradients at 298 K. The transverse relaxation rates (Rexeff) of PhoBNF20D were obtained with frequency of 40, 80, 120, 320, 480, 640, 800 and 960 Hz.
All NMR spectra were processed using NMRpipe[19] and analyzed by NMRView[20]. The data of CPMG relaxation dispersion data were fitting by NESSY[21].
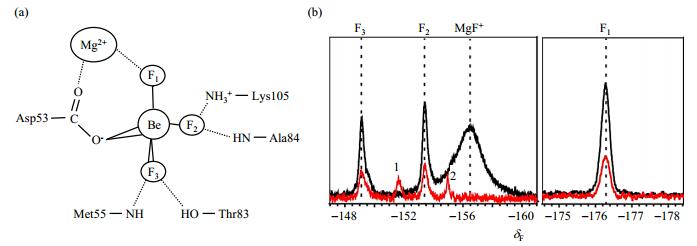
2 Results and discussion 2.1 Partially BeF3--bound PhoBNF20D observed in the absence of Mg2+In recent years, BeF3--bound PhoBNF20D observed has facilitated numerous structural studies of protein in their phosphorylated forms[22-24]. For most of the RRs, the binding of BeF3--bound PhoBNF20D observed is Mg2+-dependent. The crystal structure of BeF3--bound RR468, for instance, gives a clear picture of the interaction network [Fig. 1(a)]. Mg2+ plays an important role in the interaction between Asp53 and BeF3-. However, the BeF3- titration experiments of PhoBNF20D indicated that they can bind BeF3- with or without Mg2+ [Fig. 1(b)]. The three fluorine atoms in 19F NMR spectra [marked as F1, F2, and F3 in Fig. 1] are the indicators of BeF3- binding and have been assigned as described in reference[17]. The intensities of these three 19F NMR signals are significantly weak in the spectrum without Mg2+ compared with those in the presence of Mg2+, suggesting that only a small amount of phosphorylated conformation can be formed in the absence of Mg2+. Another two redundant 19F NMR signals which are labeled with 1 and 2 have shown up in Fig. 1(b). They may be the signals of the nonspecific binding between PhoBNF20D and BeF3-. Analysis of the crystal structure revealed that there are many charged residues near the phosphorylation site of PhoBN. Be2+ and F- can form BeF+ or BeF42- in solution, in addition to BeF3-. We speculate that these ions may interact with those charged residues. The intensity of peak 2 decreases when BeF3- increases. When Mg2+ was added, peak 1 and 2 disappeared. It indicates that Mg2+ participates in the coordination of BeF3- and PhoBNF20D, and helps BeF3- to adopt the coordination mode that is illustrated in Fig. 1(a).
|
Fig. 1 19F spectra of PhoBNF20D. (a) Schematic diagram of the interactions between BeF3- and PhoBNF20D; (b) 19F NMR spectra of 0.3 mmol/L PhoBNF20D with 1.5 mmol/L BeF3- in the presence of 10 mmol/L Mg2+ (black) or absence of Mg2+ (red) |
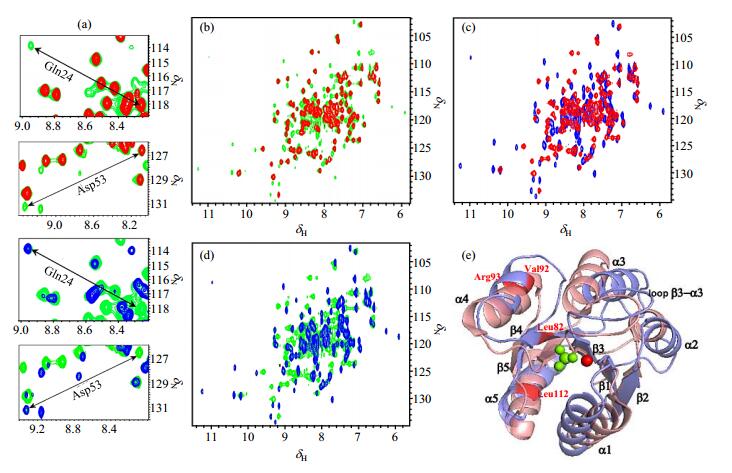
The 1H-15N HSQC spectrum of PhoBNF20D in the absence of Mg2+ shows that the protein can partially bind BeF3-, which is consistent with the 19F NMR spectrum [Fig. 1(b) and Fig. 2(b)]. Another two sets of signals appear in the Mg2+-absent spectra [green signals in Fig. 2(b) and 2(d)]. They are the spectra of apo PhoBNF20D [red signals in Fig. 2(b) and 2(c)] and BeF3--PhoBNF20D in the presence of Mg2+ [blue signals in Fig. 2(c) and 2(d)]. The enlarged spectra in Fig. 2(a) show the examples of Gln24 and Asp53 at different conditions. We also verified that the orders of addition of Mg2+ or BeF3- have no effects on the results of the spectra. The crystal structures of PhoBN in inactive and active states have been compared in Fig. 2(e). The conformational differences between two conformations mainly locate in two helical regions, helix α3 and helix α4. The helix α3 is slanted outward at the C-terminus of helix in the active conformation with respect to that in the inactive conformation. The helix α4 is longer in the active conformation than that in the inactive conformation.
|
Fig. 2 The overlay of 2D 1H-15N HSQC spectra of PhoBNF20D in different conditions. (a) The enlarged spectra of residues Gln24 and Asp53. Color codes are the same as (b), (c) and (d), respectively; (b) apo PhoBNF20D (red) and PhoBNF20D with BeF3- in the absence of Mg2+ (green); (c) apo PhoBNF20D (red) and BeF3- -PhoBNF20Din the presence of Mg2+ (blue); (d) BeF3--PhoBNF20D in the presence of (blue) or absence of (green) Mg2+; (e) Cartoon representative of the superimposed PhoBN in the inactive (slate) and active (salmon) states (PDB code: 1B00 and 1ZES), BeF3- is colored in green. Mg2+ is shown in red sphere, and the residues mentioned in Fig. 4 are colored in red and indicated by red letters |
|
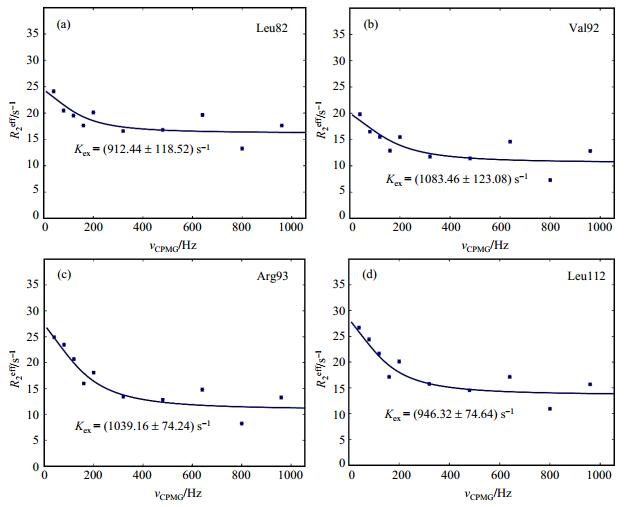
Fig. 4 15N CPMG relaxation dispersion profiles for the apo PhoBNF20D. The data were fitted globally to the fast form of the Carver-Richards equation. The data of residues (a) Leu82, (b) Val92, (c) Arg93, and (d) Leu112 are shown with the fitting curves and the Kex values |
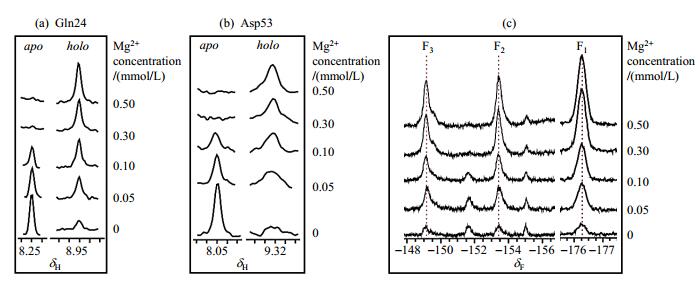
Comparison of 1H-15N HSQC spectra indicates that PhoBNF20D has pre-active conformation which is stabilized by BeF3- (Fig. 2). We choose some residues in the Mg2+-absent spectrum (green signals in Fig. 2) and calculate their intensities. The ratio of the two sets of signals in Fig. 2(b) indicates that there are approximately 15~20% pre-active conformation of apo PhoBNF20D. This pre-active conformation could completely transfer to active conformation when Mg2+ is added. The slices of Gln24 and Asp53 from HSQC titration spectra were taken as examples. As shown in Fig. 3(a) and 3(b), the signals of inactive conformation are gradually decreased, and the signals of active conformation are gradually enhanced with the increase of Mg2+ concentration. Accordingly, the signals of BeF3--PhoBNF20D in 19F NMR spectra increase while the Mg2+ concentration increases [Fig. 3(c)]. These phenomena indicate that Mg2+ plays an essential role in conformational exchanges of PhoBNF20D.
|
Fig. 3 1H-15N HSQC and 19F NMR spectra in Mg2+ titration experiments of PhoBNF20D in the presence of BeF3- (PhoBNF20D:BeF3-=1:5), the holo state refers to the active conformation of PhoBNF20D. (a) 1D slices of residue Gln24 in 1H-15N HSQC spectra; (b) 1D slices of residue Asp53 in 1H-15N HSQC spectra; (c) 19F NMR spectra of BeF3--PhoBNF20D |
15N NMR CPMG relaxation dispersion experiments were originally described by Loria et al.[25] and Vallurupalli et al.[26] in combination with an independently determined exchange-free transverse relaxation rate[27]. For a resonance undergoing exchange, the transverse relaxation rate (R2eff) has been extensively used to quantify conformational exchange processes in the millisecond to high microsecond time regime in CPMG experiment. The conformational exchanges of apo PhoBNF20D were determined by CPMG experiments. The CPMG relaxation dispersion profiles of PhoBNF20D are shown in Fig. 4. We discovered that R2eff values of a number of residues have exchanged, such as Leu82, Val92, Arg93 and Leu112. These residues are mainly located in helix α4, and it is known that the conformation of helix α4 differs in the inactive and the active conformation of PhoBN from its crystal structures (1B00 and 1ZES). The CPMG data of these residues are globally fitted and they give similar exchange constants of Rexeff (Fig. 4). We only determined the exchange rates of residues in helix α4. The exchange rates of residues in helix α3 can not be calculated due to their poor signal to noise ratio.
It is well established that conformational dynamics plays a critical role in protein function, particularly in signal transduction and regulation which typically utilizes an inactive to active state conformational transition as a means of signaling[28-30]. Molecular dynamics (MD) simulations have been used to investigate the process of RR conformational transition with atomic details which is difficult to be obtained by experimental methods[31, 32]. We directly visualized the pre-active conformation of apo PhoBNF20D by 19F NMR. These results reveal that pre-active conformation has a low population in the absence of phosphorylation and Mg2+ binding for PhoBNF20D which supports a "population shift" model for PhoBNF20D activation. Moreover, our results also indicate that Mg2+ binding can shift conformational transition towards active form.
3 ConclusionIn summary, apo PhoBNF20D has pre-active conformation in solution which can be stabilized by BeF3-, and Mg2+ accelerates this process and makes PhoBNF20D become active conformation. Our results provide basic information for better understanding of the phosphorylation process of response regulator.
| [1] | HOCH J A. Two-component and phosphorelay signal transduction[J]. Curr Opin Microbiol, 2000, 3(2): 165-170. DOI: 10.1016/S1369-5274(00)00070-9. |
| [2] | CHANG C, STEWART R C. The two-component system regulation of diverse signaling pathways in prokaryotes and eukaryotes[J]. Plant Physiol, 1998, 117(3): 723-731. DOI: 10.1104/pp.117.3.723. |
| [3] | ZSCHIEDRICH C P, KEIDEL V, SZURMANT H. Molecular mechanisms of two-component signal transduction[J]. J Mol Biol, 2016, 428(19): 3752-3775. DOI: 10.1016/j.jmb.2016.08.003. |
| [4] | GOULIAN M. Two-component signaling circuit structure and properties[J]. Curr Opin Microbiol, 2010, 13(2): 184-189. DOI: 10.1016/j.mib.2010.01.009. |
| [5] |
WANG D, LIU Y X, KOU X H, et al. NMR studies on key residues that affect phosphorylation and dephosphorylation processes of bacterial response regulator RR468[J].
Chinese J Magn Reson, 2017, 34(4): 397-407.
王丹, 刘乙祥, 寇新慧, 等. 细菌反应调节蛋白RR468磷酸化和去磷酸化关键位点的NMR研究[J]. 波谱学杂志, 2017, 34(4): 397-407. |
| [6] | NEEDHAM J V, CHEN T Y, FALKE J J. Novel ion specificity of a carboxylate cluster magnesium(Ⅱ) binding site: Strong charge selectivity and weak size selectivity[J]. Biochemistry, 1993, 32(13): 3363-3367. DOI: 10.1021/bi00064a020. |
| [7] |
LIU T, LIU M L, JIANG L. NMR analysis of divalent metalsbinding to the responseregulator YycF[J].
Chinese J Magn Reson, 2016, 33(1): 77-88.
刘婷, 刘买利, 姜凌. 二价金属离子与YycFN相互作用的NMR研究[J]. 波谱学杂志, 2016, 33(1): 77-88. |
| [8] | KOJETIN D J, THOMPSON R J, BENSON L M, et al. Structural analysis of divalent metals binding to the Bacillus subtilis response regulator Spo0F: the possibility for in vitro metalloregulation in the initiation of sporulation[J]. BioMetals, 2005, 18(5): 449-466. DOI: 10.1007/s10534-005-4303-8. |
| [9] | VOLKMAN B F, KERN D. Two-state allosteric behavior in a single-domain signaling protein[J]. Science, 2001, 291(5512): 2429-2433. DOI: 10.1126/science.291.5512.2429. |
| [10] | LAMARCHE M G, WANNER B L, CREPIN S, et al. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis[J]. FEMS Microbiol Rev, 2008, 32(3): 461-473. DOI: 10.1111/j.1574-6976.2008.00101.x. |
| [11] | GAO R, STOCK A M. Quantitative kinetic analyses of shutting off a two-component system[J]. MBio, 2017, 8(3): e00412-17. |
| [12] | SOL M, GOMISR TH F X, SERRANO L, et al. Three-dimensional crystal structure of the transcription factor PhoB receiver domain[J]. J Mol Biol, 1999, 285(2): 675. DOI: 10.1006/jmbi.1998.2326. |
| [13] | BACHHAWAT P, SWAPNA G V, MONTELIONE G T, et al. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states[J]. Structure, 2005, 13(9): 1353-1363. DOI: 10.1016/j.str.2005.06.006. |
| [14] | CANALS A, BLANCO A G, COLL M. Sigma70 and PhoB activator: getting a better grip[J]. Transcription, 2012, 3(4): 160-164. DOI: 10.4161/trns.20444. |
| [15] | MACK T R, GAO R, STOCK A M. Probing the roles of the two different dimers mediated by the receiver domain of the response regulator PhoB[J]. J Mol Biol, 2009, 389(2): 349-364. DOI: 10.1016/j.jmb.2009.04.014. |
| [16] | KOU X H, LIU X H, LIU Y X, et al. Backbone resonance assignment of the response regulator protein PhoBNF20D from Escherichia coli[J]. Biomol NMR Assign, 2018, 12(1): 133-137. DOI: 10.1007/s12104-017-9795-y. |
| [17] | LIU Y X, ROSE J, HUANG S J, et al. A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases[J]. Nat Commun, 2017, 8: 2104. DOI: 10.1038/s41467-017-02310-9. |
| [18] | CREAGER-ALLEN R L, SILVERSMITH R E, BOURRET R B. A link between dimerization and autophosphorylation of the response regulator PhoB[J]. J Biol Chem, 2013, 288(30): 21755-21769. DOI: 10.1074/jbc.M113.471763. |
| [19] | DELAGLIO F, GRZESIEK S, VUISTER G W, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes[J]. J Biomol NMR, 1995, 6(3): 277-293. |
| [20] | JOHNSON B A. Using NMRView to visualize and analyze the NMR spectra of macromolecules[M]//KRISTINA D A. Protein NMR Techniques. 2004, 278: 313-352. |
| [21] | BIERI M, GOOLEY P R. Automated NMR relaxation dispersion data analysis using NESSY[J]. BMC Bioinformatics, 2011, 12: 421. DOI: 10.1186/1471-2105-12-421. |
| [22] | CHO H, WANG W R, KIM R, et al. BeF3- acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF3- complex with phosphoserine phosphatase[J]. Proc Natl Acad Sci U S A, 2001, 98(15): 8525-8530. DOI: 10.1073/pnas.131213698. |
| [23] | WEMMER D E, KERN D. Beryllofluoride binding mimics phosphorylation of aspartate in response regulators[J]. J Bacteriol, 2005, 187(24): 8229-8230. DOI: 10.1128/JB.187.24.8229-8230.2005. |
| [24] | LIU Y X, MAO X A, LIU M L, et al. Beryllium fluoride exchange rate accelerated by Mg2+as discovered by 19F NMR[J]. J Phys Chem A, 2015, 119(1): 24-28. |
| [25] | LORIA J P, RANCE M, PALMER III A G. A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy[J]. J Am Chem Soc, 1999, 121(10): 2331-2332. DOI: 10.1021/ja983961a. |
| [26] | VALLURUPALLI P, HANSEN D F, STOLLAR E, et al. Measurement of bond vector orientations in invisible excited states of proteins[J]. Proc Natl Acad Sci U S A, 2007, 104(47): 18473-18477. DOI: 10.1073/pnas.0708296104. |
| [27] | GARDINO A K, KERN D. [5] - Functional dynamics of response regulators using NMR relaxation techniques[M]//SIMON M I, CRANE B R, CRANE A Eds. Methods Enzymol. Academic Press, 2007: 149-165. |
| [28] | KERN D, ZUIDERWEG E R P. The role of dynamics in allosteric regulation[J]. Curr Opin Struct Biol, 2003, 13(6): 748-757. DOI: 10.1016/j.sbi.2003.10.008. |
| [29] | KARPLUS M, KURIYAN J. Molecular dynamics and protein function[J]. Proc Natl Acad Sci U S A, 2005, 102(19): 6679-6685. DOI: 10.1073/pnas.0408930102. |
| [30] | HENZLER-WILDMAN K, KERN D. Dynamic personalities of proteins[J]. Nature, 2007, 450(7172): 964-972. DOI: 10.1038/nature06522. |
| [31] | FORMANECK M S, MA L, CUI Q. Reconciling the "old" and "new" views of protein allostery: a molecular simulation study of chemotaxis Y protein (CheY)[J]. Proteins, 2006, 63(4): 846-867. DOI: 10.1002/prot.20893. |
| [32] | HU X H, WANG Y M. Molecular dynamic simulations of the N-terminal receiver domain of NtrC reveal intrinsic conformational flexibility in the inactive state[J]. J Biomol Struct Dyn, 2006, 23(5): 509-517. DOI: 10.1080/07391102.2006.10507075. |
 2019, Vol. 36
2019, Vol. 36 
