2. Suzhou Institute of Nano-Tech and Nano-Bionics, Chinese Academy of Sciences, Suzhou 215123, China
2. 中国科学院 苏州纳米技术与纳米仿生研究所, 江苏 苏州 215123
Stem cell transplantation is a potential treatment option to regenerate tissue or organ functions that are impaired by acute injury or chronic degenerate diseases. In this respect, in vivo tracking of the cell transplants is critical to obtain information on their in vivo fates[1]. Many in vivo imaging approaches have been investigated for potential application in in vivo cell tracking, including magnetic resonance imaging (MRI)[2-4], positron emission tomography (PET)[5, 6], and optical imaging[7, 8], etc. MRI is regarded as one of the most powerful tools for cell tracking due to its deep tissue penetration depth, high spatial resolution, and non-invasive nature. Optical imaging is inexpensive, sensitive, and provides excellent temporal resolution, but it is limited to a tissue penetration depth of a few millimeters. These two imaging approaches are thus complementary in many aspects.
Exogenous stem cells are usually labeled with a specific imaging probe in order to be distinguished from its host tissue. A superparamagnetic agent such as iron oxide nanoparticles[9, 10] or a paramagnetic agent such as Gd chelates[11, 12] is usually used as an MRI contrast agent (CA). Iron oxide nanoparticles are usually used as T2 CA. Gd chelates are usually used as T1 CA. Quantum dots[7], fluorescent proteins[8], or fluorescent small molecules[13-15] are usually used as optical imaging probes.
Recently, we have reported that[16-19] labeling of human mesenchymal stem cells (hMSCs) with Gd agents via electroporation (EP) can induce cell-assembly of the Gd agents into nanoclusters in the cytoplasm. Along with the simultaneously introduced free Gd agents, a T1/T2 dual-mode MR imaging strategy was developed for in vivo tracking of stem cell transplants. The strategy reveals abundant information on in vivo fates of the cells. However, Gd agents have long been used as T1 contrast agents. Their T2 performance, particularly the T1/T2 dual-mode performance, needs to be cross examined by other imaging approaches.
For this purpose, we designed and synthesized a Gd-based MR-fluorescent dual-mode imaging probe. Although MR-fluorescent imaging probe have been reported which usually used traditional fluorescent dye as fluorescent group[13-15], we decided to couple Gd-DOTA with a simple small fluorescent molecule guaiazulene, 1, 4-dimethyl-7-isopropyl azulene (GA). GA is a dark blue extract from gorgonian acanthogorgia species with high yield[20], and was selected as our fluorescent probe because its color can be easily changed via chemical functionalization[21, 22]. Here we report our preliminary results on this work.
1 Materials and methods 1.1 Materials1, 4, 7, 10-tetraazacyclododecane hydrochloride was purchased from Shanghai Titan Scientific Co. Ltd. (China). GA was purchased from Jiangxi East Flavor & Fragrance Co. Ltd. (China). Other chemicals were purchased from Sinopharm Chemical Reagent Company (China). All chemicals are of analytical grade. Cell viability assay kit (MTT) was purchased from Beyotime (China). Fetal bovine serum (FBS), DMEM-F12 medium, penicillin-streptomycin, pancreatin, and all other cell culture related reagents were purchased from Gibco (USA). Milli-Q water (18.2 MΩ·cm) was used throughout the experiments and the system was purchased from Merck Millipore (China).
1.2 Synthesis of Gd-DOTA-PEG-GA1-(acetic acid)-4, 7, 10-tris(tert-butoxycarbonyl methyl)-1, 4, 7, 10-tetraazacyclodo-decane [DOTA(OtBu)3] was synthesized from hydrochloride with a multistep procedure as described in literature[23]. 2-(5-Isopropyl-3, 8-dimethylazulen-1-yl)-2-oxoacetic acid (GA-C2HO3) was synthesized by chloroacetylation of GA as described by Wang et al.[24]
Fig. 1 illustrates the specific coupling process of DOTA and GA. The final DOTA-PEG-GA was purified by high performance liquid chromatography (HPLC) with purity > 95%. Gadolinium chelating of DOTA-PEG-GA was conducted according to a previously reported procedure[16, 17]. The molecular weight (MW) of the target product Gd-DOTA-PEG-GA was measured by electrospray ionization-mass spectrometry (ESI-MS, Agilent 1200/6220) for structural confirmation.
|
Fig. 1 Synthesis process of Gd-DOTA-PEG-GA |
To investigate the fluorescent characteristic of Gd-DOTA-PEG-GA, fluorescence spectroscopy was performed with Gd-DOTA-PEG-GA dissolved in ultrapure water at concentration of 10 μmol/L in a 3 mL quartz cuvette of 1 cm path length. In addition, time-variations of fluorescence emission spectra of Gd-DOTA-PEG-GA during 0~7 days were also measured to determine its stability.
1.4 Relaxivity of aqueous Gd-DOTA-PEG-GAMRI scans of aqueous Gd-DOTA-PEG-GA solutions were performed on a Bruker AVANCE 500WB spectrometer (Bruker Biospin, Germany) with an 89 mm vertical bore magnet of 11.7 T using a birdcage coil with i.d. of 15 mm at probe temperature of about 22 ℃. Gd-DOTA was used as a control throughout this work. Both T1- and T2-relaxation times with different Gd concentrations were measured. T1- and T2-relaxation rates were characterized by the reciprocal of T1- and T2-relaxation times, respectively. Gd concentration of the solutions was measured by inductively coupled plasma-mass spectrometry (ICP-MS, Thermo X Series2, USA). Aqueous r1 and r2 in unit of mmol-1·L·s-1 were calculated through curve-fitting of T1- and T2-relaxation rates versus Gd concentrations (mol/L).
T1-relaxation time was acquired by using RARE sequence with echo time (TE) = 7 ms; repetition time (TR) = 40, 70, 100, 180, 300, 500, 750, 1 000, 1 500, 3 000, 5 000 ms; field of view (FOV) = 12×12 mm2; matrix = 96×96; slice thickness/gap = 0.8/0.2 mm; number of average = 2. T2-relaxation time was acquired by using multi slice multi echo (MSME) sequence with TR=3 000 ms, TE=8~480 ms, FOV=12×12 mm2, matrix=96×96, slice thickness/gap = 0.8/0.2 mm, and number of average = 2.
1.5 EP-labeling of hMSCs with Gd-DOTA-PEG-GAhMSCs were seeded into 100 mm × 20 mm style cell culture dishes at a density of about 1×106 cells per dish and maintained for 24 h at 37 ℃. Cells were trypsinized and centrifuged at 1 000 rpm for 5 min. The precipitated cells were resuspended in 200 µL EP-buffer in the presence of Gd-DOTA-PEG-GA at probe concentrations between 0~10 mmol/L, and were transferred to 96-well plates, respectively. Six electrical pulses of 100 μs at about 120 V and an interval of 1 s were then applied to the cells using X-Porator® EBXP-H1 (Etta Biotech, China). After EP-labeling, the cells were collected and suspended in 4 mL DMEM-F12, and allowed to recover for 15 min.
1.6 MTT cytotoxicity assayCell viability of hMSCs after EP-labeling with Gd-DOTA-PEG-GA was assessed by MTT assay. hMSCs were suspended in 200 μL EP-buffer containing 0, 1, 2, 5 and 10 mmol/L of Gd-DOTA-PEG-GA, respectively. Defined electrical pulses were applied to the cells. hMSCs without any probe labeled were treated as control. After EP-labeling, the cells were collected and suspended in 4 mL DMEM-F12, rinsed twice with 4~6 mL PBS, and transferred into a 96-well plate (8×103 cells per well). In vitro cell viability of Gd-DOTA-PEG-GA labeled hMSCs was assessed by using a standard MTT cytotoxicity assay. Each experiment was repeated five times. Statistical significance was evaluated using t‑test ANOVA analysis. p < 0.05 was considered to indicate a statistically significant difference.
1.7 TEM of hMSCs labeled with Gd-DOTA-PEG-GACellular transmission electron microscopy (TEM) was performed to precisely localize the Gd-DOTA-PEG-GA distribution in hMSCs. Labeled hMSCs were transferred into a microcentrifuge tube and spun at 1 000 rpm for 5 min. The supernatant was removed, and 1 mL 2.5% glutaraldehyde buffered with PBS was added into the microcentrifuge tube to fix hMSCs at 4 ℃ overnight. Afterwards, the cell mass was embedded in 5% agar gel and fixed in 2.5% glutaraldehyde buffered with PBS at 4 ℃ at least for 6 h followed by washing with PBS twice. Cells were then fixed in 1% osmium tetraoxide buffered with PBS for 1 h and washed three times with PBS. The samples were dehydrated using a series of acetone treatment (30%, 50%, 70%, 80%, 90%, 3×100%) of 15 min each, followed by embedded in 1:1 epoxy resin/acetone solution for 1 h. The samples were then transferred into capsules containing fresh 100% epoxy resin and fixed for at least 2 h. The capsules were then left in a furnace at a temperature of 70 ℃ for 2 days to polymerize the epoxy resin. After quickly cooling down, the hardened samples were cut into 70~90 nm thick sections with Ultramicrotome (Leica UC6, Austria) and applied on a copper grid for TEM observation (Tecnai G2 F20 S-Twin, FEI, USA).
1.8 In vitro MRI of hMSCs labeled with Gd-DOTA-PEG-GAhMSCs labeled with Gd-DOTA or Gd-DOTA-PEG-GA at concentrations of 0, 2, 5, and 10 mmol/L were seeded in 100 mm × 20 mm dishes (about 1×106 cells/dish), respectively. When cell density growing to 90%, half of EP-labeled hMSCs were used for proliferation and the other half were treated for in vitro MRI. For MRI, hMSCs were trypsinized, centrifuged, and washed twice with PBS. The harvested cells were transferred into a capillary with i.d. of about 1.0 mm and packed into cell pellets by centrifugation at 1 200 rpm for 5 min. T1- and T2-weighted images of hMSC pellets were collected on the 11.7 T MRI system. T1- and T2-relaxation times were also measured.
T1-weighted images were acquired by using MSME sequence with TE = 5.2 ms, TR = 500 ms, FOV = 12×12 mm2, matrix = 96×96, slice thickness/gap = 0.8/0.2 mm, and number of average = 4. T2-weighted images were acquired by using MSME sequence with TE = 40 ms, TR = 3 000 ms, FOV = 12×12 mm2, matrix = 96×96, slice thickness/gap = 0.8/0.2 mm, and number of average = 2.
1.9 In vitro fluorescence imaging of EP-labeled hMSCsFor in vitro tracking of hMSCs by fluorescence imaging, hMSCs were EP-labeled with Gd-DOTA-PEG-GA at a concentration of 5 mmol/L. After EP-labeling, the cells were collected and suspended in 4 mL DMEM-F12, allowed to recover for 15 min, and then transferred into 100 mm×20 mm dishes for incubation and proliferation. After washing with PBS for 3 times, the cells were observed under an inverted microscope (Nikon Ti-E microscopy, Japan).
2 Results and discussion 2.1 MS characterization of Gd-DOTA-PEG-GAThe chemical structure of Gd-DOTA-PEG-GA was confirmed by their MW as measured by ESI-MS. Fig. 2 shows mass spectrum of Gd-DOTA-PEG-GA (C39H55GdN6O11, MW = 941.32), found at m/z 471.664 0 for [M+2H]2+.

|
Fig. 2 Mass spectrum of Gd-DOTA-PEG-GA with fragmentation energy of 120.0 V in positive mode |
Fig. 3 presents T1- and T2-relaxation rates of Gd-DOTA and Gd-DOTA-PEG-GA solutions as a function of Gd concentration. r1 and r2 of Gd-DOTA are about 3.74 and 4.85 mmol-1·L·s-1. r1 and r2 of Gd-DOTA-PEG-GA are about 5.65 and 7.74 mmol-1·L·s-1, respectively, both slightly higher than that of Gd-DOTA.

|
Fig. 3 (a) T1- and (b) T2-relaxation rates as a function of Gd concentration |
Fig. 4(a) shows the fluorescence excitation and emission spectra of Gd-DOTA-PEG-GA at concentrations of 10 μmol/L. An absorption band was observed at 425 nm and an emission band was observed at 498 nm upon excitation at a wavelength of 425 nm. Both the emission band shape and intensity remain stable during a period of 7 days [Fig. 4(b)]. The results suggest that Gd-DOTA-PEG-GA was sufficiently stable for the use as MR-fluorescence dual-mode probe.

|
Fig. 4 (a) Fluorescence emission spectrum of 10 μmol/L Gd-DOTA-PEG-GA (inset, fluorescence excitation spectrum of Gd-DOTA-PEG-GA); (b) Fluorescence emission spectra of 10 μmol/L Gd-DOTA-PEG-GA in ultrapure water during 7 days |
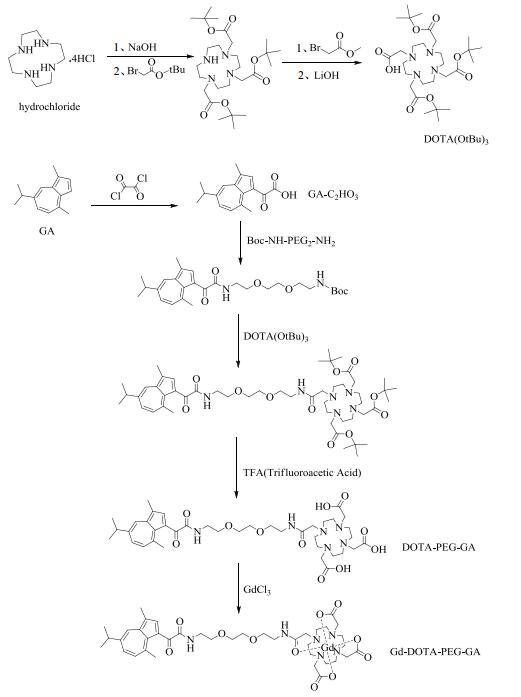
Fig. 5 illustrates possible cytotoxicity induced by either EP or the contrast agents. EP in the absence of Gd-DOTA-PEG-GA (EP0) exerted minor adverse effects on the survival of hMSCs with cell viability of about 95% relative to the control. EP1~EP10 in the presence of Gd-DOTA-PEG-GA at concentrations of 1, 2, 5, 10 mmol/L exerted minor additional adverse effects on hMSCs with cell viability of about 90% relative to the control. The results suggest that the contrast agent is biologically safe.
|
Fig. 5 MTT assay of hMSCs without any probe labeled (control) and EP-labeled with Gd-DOTA-PEG-GA at different concentrations (EP0: 0 mmol/L, EP1: 1 mmol/L, EP2: 2 mmol/L, EP5: 5 mmol/L, EP10: 10 mmol/L) |
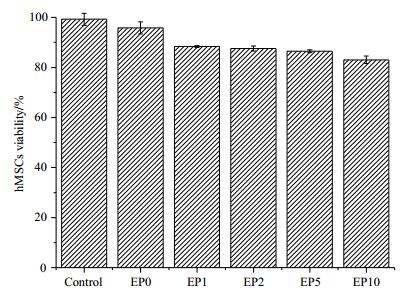
Fig. 6 presents TEM images of hMSCs EP-labeled with Gd-DOTA-PEG-GA which were subjected to fixed dehydration with 2.5% glutaraldehyde for 6 h at 4 ℃ on day 0, 1, 3 after cell labeling. EP-labeling of hMSCs induced cell-assembly of Gd-DOTA-PEG-GA nanoclusters into cytoplasm. Immediately after cell labeling (on day 0), both intracellular nanoclusters and membrane-bound Gd-DOTA-PEG-GA were observed [Fig. 6(a)]. In addition, we also noted that significant amount of free Gd-DOTA-PEG-GA agents were introduced to the cytoplasm during EP-labeling. On day 1 and day 3, membrane-bound Gd-DOTA-PEG-GA disappeared with only intracellular nanoclusters remaining in the cytoplasm [Fig. 6(b) and 6(c)]. The free Gd-DOTA-PEG-GA agents were also released quickly via exocytosis during cell culture. The findings suggest that the cell-assembled nanoclusters have longer cellular retention time than the membrane-bound Gd-DOTA-PEG-GA, in agreement with our previous findings on other MRI probes[17, 19].
|
Fig. 6 Cellular TEM images of hMSCs EP-labeled with Gd-DOTA-PEG-GA: (a) Immediately after cell labeling; (b) Followed by a 1-day culture and recovery; (c) Followed by a 3-day culture and recovery. The scale bar is 5 μm |
Fig. 7 presents T1- and T2-weighted MR images of hMSCs EP-labeled with Gd-DOTA-PEG-GA at concentrations of 2, 5, and 10 mmol/L collected immediately after cell labeling as well as subsequently after cell proliferation of defined times. A bright signal appeared in both T1- and T2-weighted images of hMSCs EP-labeled with Gd-DOTA (as a control) but persisted only about 3 days. T1-weighted images of hMSCs EP-labeled with Gd-DOTA-PEG-GA at different concentrations did not show obvious signal enhancement [Fig. 7(a)]. T2-weighted images showed a dark signal that persisted for about 7 days [Fig. 7(b)].

|
Fig. 7 In vitro (a) T1- and (b) T2-weighted MR images of hMSCs pellets EP-labeled with Gd-DOTA and Gd-DOTA-PEG-GA as a function of cell proliferation time at 11.7 T. Number below each image indicates cell proliferation time in day. (c) T1- and (d) T2-weighted MR signal intensities; (e) Cellular T1- and (f) T2-relaxation rates. MR signal intensity of dark image is tentatively given a value of 5 or below for ease of following its change profile (b). Discussion on T2 relaxation rate has to be restricted to below 50 s-1 because the accuracy of measurement of T2 relaxation rate deteriorates at above 50 s-1 (equivalent to T2 relaxation time of below 20 ms) |
The measured T1- and T2-weighted signal intensities of hMSCs are plotted as functions of cell proliferation time in Fig. 7(c) and 7(d). The measured T1- and T2-relaxation rates of hMSCs after MR image collection are also plotted as a function of cell proliferation time in Fig. 7(e) and 7(f), which can account for the measured changes in both T1- and T2-weighted signal intensities on the basis of Equ. (1)[17, 25]:
| $ F_{s e} \propto\left(1-\mathrm{e}^{-T_{\mathrm{R}} / T_{1}}\right) \mathrm{e}^{-T_{\mathrm{E}} / T_{2}} $ | (1) |
where Fse is the spin echo signal intensity, 1/T1 and 1/T2 are T1- and T2-relaxation rates, TR and TE are experimental repetition time and echo time, respectively.
Labeling of hMSCs with an MRI contrast agent accelerates both T1- and T2-relaxation rates. Free Gd-DOTA agent introduced into the cytoplasm is in favor of acceleration of T1-relaxation rate and thus MRI signal enhancement [Fig. 7(c) and 7(d), day 0 and day 1]. Subsequent fast release of free Gd-DOTA agent from the cytoplasm results in a fast recovery of T1-relaxation rate [Fig. 7(e)] associated with a fast recovery of both T1- and T2-weighted signal [Fig. 7(c) and (d), day 4 and later]. EP induced cell-assembly of Gd-DOTA-PEG-GA nanoclusters into the cytoplasm is in favor of acceleration of T2-relaxation rate and thus MRI signal reduction [Fig. 7(c) and 7(d), day 0 and day 1]. The intracellular nanoclusters have longer cellular retention time so that acceleration of T2-relaxation rate can persist over a longer period which is associated with a persistent T2-weighted signal reduction effect. The findings are also in agreement with our previous findings[17-19] on other MRI probes and thus suggest that the dual-mode probe can be used for in vivo MRI tracking of transplanted stem cells.
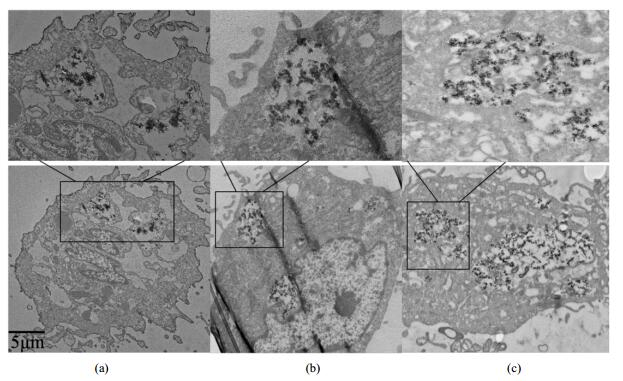
2.7 Fluorescence imagingFig. 8 presents fluorescence images of hMSCs EP-labeled with Gd-DOTA-PEG-GA at a concentration of 5 mmol/L under an inverted microscope. Bright field image confirmed that the cells were viable throughout the experiments after EP-labeling with Gd-DOTA-PEG-GA [Fig. 8(a)]. The nanoclusters were distributed within the cytoplasm, emitting green fluorescence under UV excitation [Fig. 8(b)]. Overlay of bright and dark field images demonstrates that the green fluorescence was from both the cell membrane and the intracellular region [Fig. 8(c)]. The results indicate the potential of Gd-DOTA-PEG-GA as a fluorescent nanoprobe.
|
Fig. 8 Fluorescence images of hMSCs EP-labeled with Gd-DOTA-PEG-GA at a concentration of 5 mmol/L under inverted microscope: (a) Bright field; (b) Dark field; (c) Overlay of (a) and (b). The scale bar is 50 μm |
Combining with its MRI performance, it can function as a MR-fluorescent dual-mode probe for stem cell tracking. For example, the T2 effect of this probe can be used for the in vivo MRI observation, and optic property allows the straightforward hMSCs localization for a physiological section analysis without the necessity of cell staining.
3 ConclusionGd-DOTA was coupled with GA to yield an MR-fluorescent dual-mode imaging probe for stem cell tracking. The probe was used to label hMSCs via electroporation. EP-labeling induced cell-assembly of the probe into nanoclusters in the cytoplasm as confirmed by cellular TEM. Cellular MRI of EP-labeled hMSCs demonstrated a significant T2-weighted signal reduction effect which persisted over 7 days. The probe has an UV absorption band at 425 nm, and a fluorescent band at 498 nm with good stability. hMSCs labeled with the probe exhibited a green fluorescence under inverted microscope. The biosafety was preliminarily assessed by MTT assay which yielded a high cellular viability for EP-labeling with the probe. These characteristics allow the probe to function as an MR-fluorescent dual-mode imaging probe for cell tracking.
Acknowledgement The authors acknowledge Karebay Biochem Inc. for assistance with synthesis of DOTA-PEG-GA.| [1] | GERA A, STEINBERG G K, GUZMAN R. In vivo neural stem cell imaging: current modalities and future directions[J]. Regen Med, 2010, 5(1): 73-86. DOI: 10.2217/rme.09.79. |
| [2] | KRAITCHMAN D L, BULTE J W. Imaging of stem cells using MRI[J]. Basic Res Cardiol, 2008, 103(2): 105-113. DOI: 10.1007/s00395-008-0704-5. |
| [3] | POLITI L S. MR-based imaging of neural stem cells[J]. Neuroradiology, 2007, 49(6): 523-534. DOI: 10.1007/s00234-007-0219-z. |
| [4] | ROGERS W J, MEYER C H, KRAMER C M. Technology insight: in vivo cell tracking by use of MRI[J]. Nat Rev Cardiol, 2006, 3(10): 554-562. DOI: 10.1038/ncpcardio0659. |
| [5] | ZHANG H, SONG F H, XU C Y, et al. Spatiotemporal PET imaging of dynamic metabolic changes after therapeutic approaches of induced pluripotent stem cells, neuronal stem cells, and a Chinese patent medicine in stroke[J]. J Nucl Med, 2015, 56(11): 1774-1779. DOI: 10.2967/jnumed.115.163170. |
| [6] | WANG J C, CHAO F F, HAN F, et al. PET demonstrates functional recovery after transplantation of induced pluripotent stem cells in a rat model of cerebral ischemic injury[J]. J Nucl Med, 2013, 54(5): 785-792. DOI: 10.2967/jnumed.112.111112. |
| [7] | CHEN G C, TIAN F, LI C Y, et al. In vivo real-time visualization of mesenchymal stem cells tropism for cutaneous regeneration using NIR-II fluorescence imaging[J]. Biomaterials, 2015(53): 265-273. |
| [8] | CHENG G C, LIN S Y, HUANG D H, et al. Revealing the fate of transplanted stem cells in vivo with a novel optical imaging strategy[J]. Small, 2018, 14(3). DOI: 10.1002/smll.201702679. |
| [9] | MAHMOUDI M, HOSSEINKHANI H, HOSSEINKHANI M, et al. Magnetic resonance imaging tracking of stem cells in vivo using iron oxide nanoparticles as a tool for the advancement of clinical regenerative medicine[J]. Chem Rev, 2011, 111(2): 253-280. DOI: 10.1021/cr1001832. |
| [10] |
ZHANG H Y, LI C L, YING X F, et al. A gadolinium-based T1 MRI probe for detection of lung cancer stem cells[J].
Chinese J Magn Reson, 2016, 33(4): 627-634.
张宏岩, 李春林, 英晓芳, 等. 靶向肺癌干细胞的多肽Gd基T1型MRI探针的研究[J]. 波谱学杂志, 2016, 33(4): 627-634. |
| [11] | AGUDELO C A, TACHIBANA Y, HURTADO A F, et al. The use of magnetic resonance cell tracking to monitor endothelial progenitor cells in a rat hindlimb ischemic model[J]. Biomaterials, 2012, 33(8): 2439-2448. DOI: 10.1016/j.biomaterials.2011.11.075. |
| [12] | TACHIBANA Y, ENMI J I, AGUDELO C A, et al. Long-term/ bioinert labeling of rat mesenchymal stem cells with PVA-Gd conjugates and MRI monitoring of the labeled cell survival after intramuscular transplantation[J]. Bioconjugate Chem, 2014, 25(7): 1243-1251. DOI: 10.1021/bc400463t. |
| [13] | HUBER M M, STAUBLI A B, KUSTEDJO K, et al. Fluorescently detectable magnetic resonance imaging agents[J]. Bioconjugate Chem, 1998, 9(2): 242-249. DOI: 10.1021/bc970153k. |
| [14] | KIM E J, BHUNIYA S, LEE H, et al. In vivo tracking of phagocytic immune cells using a dual imaging probe with gadolinium-enhanced MRI and near-infrared fluorescence[J]. ACS Appl Mater Interfaces, 2016, 8(16): 10266. DOI: 10.1021/acsami.6b03344. |
| [15] | ZHU T F, MA X Q, CHEN R H, et al. Using fluorescently-labeled magnetic nanocomposites as a dual contrast agent for optical and magnetic resonance imaging[J]. Biomater Sci, 2017, 5(6): 1090-1100. DOI: 10.1039/C7BM00031F. |
| [16] | CAO L M, LI B B, YI P W, et al. The interplay of T1- and T2-relaxiation on T1-weighted MRI of hMSCs induced by Gd-DOTA-peptides[J]. Biomaterials, 2014, 35(13): 4168-4174. DOI: 10.1016/j.biomaterials.2014.01.073. |
| [17] | ZHANG Y H, ZHANG H Y, LI B B, et al. Cell-assembled (Gd-DOTA)i -triphenylphosphonium (TPP) nanoclusters as a T2 contrast agent reveal in vivo fates of stem cell transplants[J]. Nano Res, 2018, 11(3): 1625-1641. DOI: 10.1007/s12274-017-1778-x. |
| [18] |
ZHANG Y H, ZHANG H Y, ZHANG H L, et al. Preparation and application of a new-type Gd agents as T2 contrast agent[J].
Chinese J Magn Reson, 2017, 34(3): 302-310.
张艳辉, 张宏岩, 张海禄, 等. 新型Gd基T2造影剂的制备和应用[J]. 波谱学杂志, 2017, 34(3): 302-310. |
| [19] | ZHANG P L, ZHANG Y H, LI B B, et al. Cell-assembled nanoclusters of MSC-targeting Gd-DOTA-peptide as a T2 contrast agent for MRI cell tracking[J]. J Pept Sci, 2018, 24(4/5): e3077. |
| [20] | CHEN D W, YU S J, OFWEGEN L V, et al. Anthogorgienes A-O, new guaiazilene-derived terpenoids from a Chinese gorgonian anthogorgia species, and their antifouling and antibiotic activities[J]. J Agric Food Chem, 2012, 60(1): 112-113. DOI: 10.1021/jf2040862. |
| [21] | AMIR E, AMIR R J, CAMPOST L M, et al. Stimuli-responsive azulene-based conjugated oligomers with polyaniline-like properties[J]. J Am Chem Soc, 2011, 133(26): 10046-10049. DOI: 10.1021/ja203267g. |
| [22] | DONG J X, ZHANG H L. Azulene-based organic function molecules for optoelectronics[J]. Chinese Chem Lett, 2016, 27(8): 1097-1104. DOI: 10.1016/j.cclet.2016.05.005. |
| [23] | LI C, WINNARD P, BHUJWALLA Z M. Facile synthesis of 1-(acetic acid)-4, 7, 10-tris(tert-butoxycarbonylmethyl)-1, 4, 7, 10- tetraazacyclododecane: a reactive precursor chelating agent[J]. Tetrahedron Lett, 2009, 50(24): 2929-2931. DOI: 10.1016/j.tetlet.2009.03.198. |
| [24] |
WANG D L, HAN S, GU Z, et al. Synthesis of 1-(2-Benzo[b]furoyl) guaiazulene derivatives[J].
Chinese J Org Chem, 2008, 28(9): 1641-1645.
王道林, 韩珊, 谷峥, 等. 1-(2-苯并呋喃酰基)愈创兰烃薁的合成[J]. 有机化学, 2008, 28(9): 1641-1645. |
| [25] | MCROBBIE D W, MOORE E A, GRAVES M J, et al. MRI from picture to proton[M]. 2nd ed. Cambrige: Cambrige University Press, 2007. |
 2019, Vol. 36
2019, Vol. 36 
