2. National Laboratory for Clean Energy, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China;
3. Division of Virology & Immunology, National Center for AIDS/STD Control and Prevention, Beijing 102206, China;
4. State Key Laboratory of Magnetic Resonance and Atomic and Molecular Physics, National Center for Magnetic Resonance in Wuhan(Wuhan Institute of Physics and Mathematics, Chinese Academy of Sciences), Wuhan 430071, China
2. 洁净能源国家实验室(中国科学院 大连化学物理研究所), 辽宁 大连 116023;
3. 国家艾滋病性病防治中心, 病毒与免疫研究室, 北京 102206;
4. 波谱与原子分子物理国家重点实验室, 武汉磁共振中心(中国科学院 武汉物理与数学研究所), 湖北 武汉 430071
The glycine-rich RNA-binding protein, AtGrp7, was first determined as a component of a negative feedback loop in the circadian clock regulation of Arabidopsis thaliana[1]. This protein can bind to a set of transcripts such as 5′ UTR, 3′ UTR, and intron RNA in Arabidopsis plants, including its own transcript[1-3]. Through interaction with the RNA targets, AtGrp7 can promote or affect alternating splicing, pri-miRNA processing, flowering, and stomata opening and closing, besides influencing circadian oscillations at the post-transcriptional level[4-7]. AtGrp7 is also involved in RNA-based pathogen defense, cold and drought tolerance, and other stress responses[8-14]. AtGrp7 is composed of an N-terminal canonical RNA recognition motif (RRM) and a C-terminal unstructured glycine-rich domain. The RRM domain functions as an RNA chaperone in RNA metabolism, while the glycine-rich domain acts as a shuttle for RNA transport between the nucleus and cytoplasm, and contributes to RNA binding[15]. To deeply understand the biological functions of AtGrp7 in plants, high-resolution structures of AtGrp7 RRM domain, especially of its complexes with variant RNAs are necessary.
Protein preparation of RRM domains originally from eukaryotes was usually performed via a regular purification procedure. Supernatant from cell extracts was passed through an Ni-NTA column for His-tagged protein, followed by a tobacco etch virus (TEV, or other proteases) cleavage to remove the tag, and then subjected to size-exclusion chromatography[16, 17]. In some cases, an ion-exchange purification step was applied between the Ni-NTA process and size-exclusion chromatography. However, in our initial trial about the AtGrp7 RRM domain using this procedure, we observed obvious UV signals of mixed proteins and contaminants for the TEV-cleaved AtGrp7 RRM domain; the contaminants decreased following purification by size-exclusion chromatography. For this reason, other purification protocols should be tried in order to get pure AtGrp7 RRM domain for structural and biological researches.
In the present study, we aimed to obtain AtGrp7 RRM domain free of contaminants by using a two-step purification protocol of denaturing and refolding. The preliminary structure and function of the refolded AtGrp7 RRM domain were confirmed by nuclear magnetic resonance (NMR) spectroscopy and isothermal titration calorimetry (ITC).
1 Materials and methods 1.1 Cloning and overexpression of AtGrp7 RRM domain (AtGrp71-90)DNA encoding the AtGrp71-90 sequence from Arabidopsis thaliana was synthesized by Genscript, Nanjing, China. The recombinant plasmid AtGrp7RRM was obtained by inserting the synthesized DNA fragment into a modified pET16b vector at the XhoI/BamHI restriction sites. The modified pET16b was generated by insertion of the DNA fragment encoding a B1 domain of streptococcal protein G (GB1) and a TEV protease digestion site into the original pET16b vector after the N-terminal His10 tag, resulting in an extra glycine at the N-terminus of the expressed protein after TEV cleavage. The recombinant plasmid AtGrp7RRM was verified by DNA sequencing.
AtGrp71-90 was overexpressed in the Escherichia coli BL21 (DE3) strain after transformation of AtGrp7RRM. The cells were first grown in 10 mL LB broth with ampicillin (100 μg/mL) at 37 ℃ (220 rpm) until an OD600 of 0.6 was reached. For unlabeled AtGrp71-90, the cells were transferred to 1 L LB broth with ampicillin (100 μg/mL) and grown at 37 ℃ (220 rpm) until an OD600 of 0.4 was attained, and were subsequently grown at 18 ℃ and 220 rpm for 0.5~1 h. For 13C, 15N- or 15N -labeled AtGrp71-90, cells were harvested when an OD600 of 0.6 was reached, and the resulting pellet was resuspended in M9 minimum media, with 1 g/L 15N-labeled NH4Cl (Cambridge Isotope Laboratories, Andover MA) as the sole nitrogen source, and uniformly labeled 13C-glucose (2 g/L; Cambridge Isotope Laboratories, Andover MA) or unlabeled glucose (4 g/L) as the sole carbon source. Cells were grown in M9 minimum media at 37 ℃ (220 rpm), until an OD600 of 0.3 was reached, and subsequently grown at 18 ℃ and 220 rpm for 0.5~1 h. When the OD600 reached 0.6 (or 0.4) for cells grown in LB broth (or M9 minimum media) at 18 ℃, cells were induced with 0.1 mmol/L isopropyl-β-D-thiogalactoside (IPTG) at 18 ℃ and 220 rpm for 16 h. Then, the cells were centrifuged at 5 000 g for 15 min at 4 ℃, and the cell pellets were frozen in liquid nitrogen and stored at –80 ℃.
1.2 Protein refolding and purification of AtGrp71-90The cell pellets harvested from 1 L of the media were resuspended in 30 mL freshly prepared lysis buffer [50 mmol/L Tris-HCl (pH 8.0), 500 mmol/L NaCl, 8 mol/L urea, 0.5 mmol/L ethylene diamine tetraacetic acid (EDTA), 2 mmol/L dithiothreitol (DTT), 2 mmol/L phenylmethylsulfonyl fluoride (PMSF)]. The cells were sonicated at a 30% amplitude (Ningbo Scientz Biotechnology company; 15 min per cycle of 6 s on, followed by 6 s off intervals) for 5 cycles at 25 ℃, followed by shaking on an orbital shaker at 20 rpm for 1 h before centrifugation (15 000 g at 25 ℃ for 40 min). The supernatant was purified by passing through a column of Ni-NTA agarose resin pre-equilibrated with lysis buffer. After washing with high salt buffer [50 mmol/L Tris-HCl (pH 8.0), 1 mol/L NaCl, 8 mol/L urea, 0.5 mmol/L EDTA, 2 mmol/L DTT, 2 mmol/L PMSF, 20 mmol/L imidazole] and equilibration with low salt buffer (100 mmol/L NaCl; other components same as the high salt wash buffer), 40 mL elution buffer [50 mmol/L Tris-HCl (pH 8.0), 100 mmol/L NaCl, 8 mol/L urea, 0.5 mmol/L EDTA, 2 mmol/L DTT, 500 mmol/L imidazole] was used for elution, and the first 20 mL eluted fraction of the target protein was concentrated (to a final volume of 2~3 mL). The concentrated proteins were slowly added into a 40-fold volume of ice-cold refolding buffer [50 mmol/L Tris-HCl (pH 8.0), 1 mol/L NaCl, 0.5 mmol/L EDTA, 2 mmol/L DTT] drop by drop with stirring, at an interval of 30 s. The resulting solution was left overnight at 4 ℃ with stirring. The refolded proteins were concentrated to a volume of 2~3 mL again by centrifugation (2 000 g; Amicon Ultra-15, MWCO: 3 000, Millipore, USA), and then diluted with TEV buffer [50 mmol/L Tris-HCl, 100 mmol/L NaCl, 0.5 mmol/L EDTA, 5 mmol/L β-mercaptoethanol (BME), pH 8.0] to a low salt concentration (less than 200 mmol/L). Next, 18 μg/mL of TEV protease was added, and the proteins were left at 4 ℃ overnight for dialysis in TEV buffer. The TEV-cleaved protein mixture was passed through a column of Ni-NTA resin to obtain His-tagged and GB1-free samples. Then, AtGrp71-90 was loaded into a HiLoad 16/600 Superdex 75 preparation-grade column pre-equilibrated with NMR buffer [20 mmol/L NaPi (pH 6.0), 50 mmol/L NaCl, 0.5 mmol/L EDTA, 5 mmol/L BME], and purified at a flow rate of 0.5 mL/min. Finally, the AtGrp71-90 protein was exchanged with ddH2O (18.2 MΩ·cm) by dialysis, lyophilized, and stored at -20 ℃. Protein samples were dissolved in the NMR buffer for subsequent ITC and NMR experiments.
1.3 Isothermal titration calorimeter measurements6-nt (5′-TTCTGG-3′) and 32-nt (5′-ATTTTGTTCTGGTTCTGCTTTAGATTTGATGT-3′) DNA, which were DNA counterparts of the RNA from the 3′ UTR of AtGrp7[18], were synthesized by Sangon Biotech (Shanghai, China) and used without further purification. ITC experiments were performed at 25 ℃ using MicroCal ITC200 for the AtGrp71-90 protein and the 6-nt or 32-nt DNAs dissolved in the NMR buffer. The concentration of AtGrp71-90 was in a range of 0.01~0.03 mmol/L, and those of DNAs were in a range of 0.1~0.3 mmol/L. All solutions were degassed for 5 min at 25 ℃, and titrations were performed at temperatures of (25±0.1) ℃. The cell volume was 200 μL and the injection volume was 40 μL, with an interval of 120 s between 20 consecutive injections. The ITC data was analyzed by the origin 7.0 software, which was supplied by MicroCal.
1.4 NMR spectroscopyAll NMR experiments were performed on a Bruker Avance Ⅲ HD 700 MHz spectrometer equipped with a QCI cryogenic probe. Three-dimensional HNCACB, CBCA(CO)NNH, HNCA, HN(CO)CA, and HNCO spectra were recorded at 20 ℃ for the 13C, 15N-labeled proteins in free form or as complexes with 6-nt 5′-UUCUGG-3′ from 3′ UTR of AtGrp7 (synthesized by Genscript, Nanjing, China). NMR titration experiments of 15N-labeled AtGrp71-90 and 6-nt (or 32-nt) DNA were performed with protein to DNA molar ratios of 0, 1:1, and 1:2. Chemical shift differences were calculated according to the following equation:
| $ \mathit{\Delta }\delta =\sqrt{\mathit{\Delta }\delta _{\text{H}}^{2}+0.2\mathit{\Delta }\delta _{\text{N}}^{2}} $ | (1) |
All NMR data was processed using nmrPipe[19] and analyzed using nmrViewJ[20].
1.5 CS-Rosetta model generationCS-Rosetta[21-23] structural models were generated using the web server at the University of Wisconsin (https://csrosetta.bmrb.wisc.edu/csrosetta). In general, a TALOS table containing the protein sequence information and chemical shifts of 15N, 1HN, 1Hα, 13Cα, 13Cβ, and 13C′ from the NMR backbone assignment were used as the inputs, and 3 000 models were calculated. The ten lowest energy structures calculated by CS-Rosetta was chosen for structural analysis.
2 Results and discussion 2.1 Construct design, expression optimization and initial purification of AtGrp71-90The RRM domain of AtGrp7 from Arabidopsis thaliana was predicted to be 10~80 aa long, according to the Pfam database (http://pfam.xfam.org)[24]. The first 100 residues, including the predicted RRM domain, were then used to perform protein Blast from the Uniprot database (http://www.uniprot.org/blast), resulting in more than 250 RRM-containing proteins with ≥70% sequence identities, in which a three-dimensional structure of the RRM domain of Nicotiana tabacum glycine-rich RBP1 (PDB code 4C7Q; 84% sequence identity) was already resolved[25]. By sequence comparison and structure analysis, we predicted that the functional RRM domain of AtGrp7 is encompassed in the range of 7~86 aa. Finally, 1~90 N-terminal residues of AtGrp7 were selected for the present study. AtGrp71-90 was cloned into a modified pET16b vector with N-terminal His10 and GB1 (B1 domain of streptococcal protein G)-tagged, which facilitated protein purification and was propitious to the yield, solubility, and stability of target proteins[26]. A TEV cleavage site was inserted in between the DNAs encoding GB1 and AtGrp71-90 for the purpose of removing the His10-tag and GB1 in the overexpressed protein.
Protein expression of AtGrp71-90 was tested in BL21 (DE3) at 18 ℃ and 37 ℃, by induction with 1 mmol/L IPTG for 4 h, and with 0.1 mmol/L IPTG for 16 h. A little higher protein expression level was observed at 18 ℃, as indicated by SDS-PAGE [Fig. 1(a)], and this temperature was then chosen as the optimum condition for protein overexpression. However, although Dnase I was added during the lysis step, our initial purification trial of AtGrp71-90 using the regular protocol that was used for Pin1 WW domains[27] revealed that the TEV-cleaved proteins contained some contaminants, for which the UV peak centered at 269 nm indicated a mixture of proteins (~280 nm) and some contaminants like DNA or imidazole (~260 nm) [Fig. S1(b)]. The contaminant signal was not obvious after the first Ni-NTA purification, and it became clear after the second Ni-NTA step [Fig. S1(a) and (b)]. After size-exclusion chromatography, the amount of contaminants decreased. However, a shoulder peak could be implicated from the shape of the UV peak [Fig. S1(c)]. Considering the large extinction coefficients of contaminants like nucleotides or imidazole with respect to those of proteins, we explained the above phenomena as follows. Firstly, contaminants had a relatively weak binding affinity for the AtGrp7 RRM domain, and their amounts relative to those of the RRM protein were low. Secondly, the UV signal of contaminants was masked by that of overexpressed proteins with extinction coefficients ~20 000 L·mol-1·cm-1 (for GB1-fused AtGrp71-90) in the first Ni-NTA. Thirdly, after TEV cleavage, the absence of GB1 (with extinction coefficient 11 500 L·mol-1·cm-1) resulted in a greater contribution of the contaminants to the UV signal than in the first Ni-NTA step. Finally, most of the contaminants were separated from AtGrp71-90 by size-exclusion chromatography. However, a part of the contaminants remained due to their relatively weak binding to the RRM protein. Although an ion exchange before size-exclusion chromatography could improve the purity of AtGrp71-90 to some extent, the shape of the UV peak indicated that contaminants were still present in the final purified protein (data not shown).
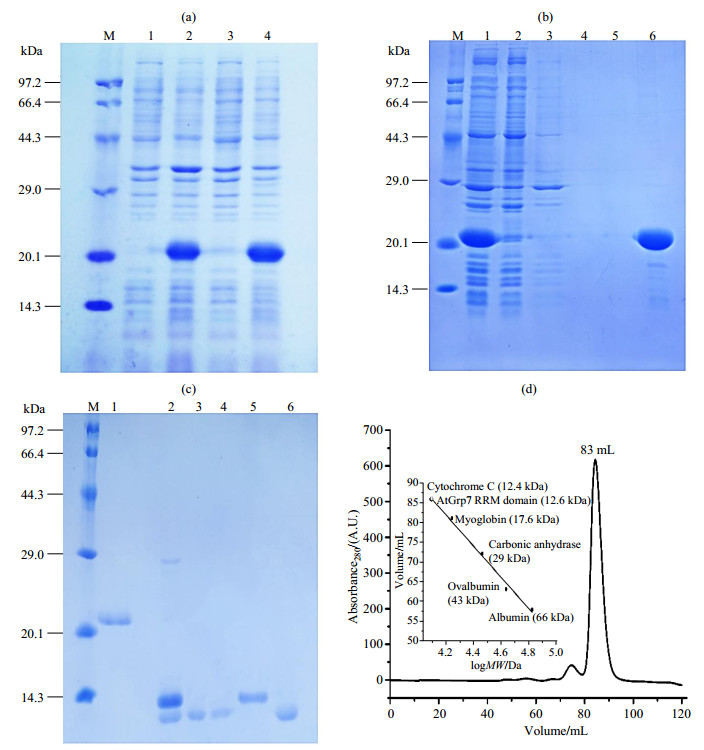
|
Fig. 1 Expression optimization and purification of AtGrp71-90. (a) 15% SDS-PAGE of the expression test of His10 and GB1-tagged AtGrp71-90 at different temperatures. M, protein marker; 1, before induction at 37 ℃; 2, induction with 1 mmol/L IPTG at 37 ℃ for 4 h; 3, before induction at 18 ℃; 4, after induction with 100 μmol/L IPTG at 18 ℃ for 16 h. (b) 15% SDS-PAGE of the first Ni-NTA purification step of His10 and GB1-tagged AtGrp71-90 in the denatured condition. M, protein marker; 1, supernatant; 2, flow through fraction; 3, high salt wash, first 50 mL; 4, high salt wash, second 50 mL; 5, low salt wash; 6, the first 20 mL of elution fraction. (c) 15% SDS-PAGE of the second Ni-NTA purification step of refolded AtGrp71-90 with His10 and GB1 tags after TEV cleavage. M, protein marker; 1, after quick-dilution refolding; 2, after TEV cleavage; 3, flow through fraction; 4, low salt (100 mmol/L NaCl; other same as TEV buffer) wash fraction; 5, elution fraction after high salt (1 mol/L NaCl) and high imidazole concentration (500 mmol/L); 6, after size exclusion chromatography. The weak band close to the marker at 29.0 kDa was from the TEV protease. (d) Size exclusion chromatography of TEV-cleaved AtGrp71-90 |
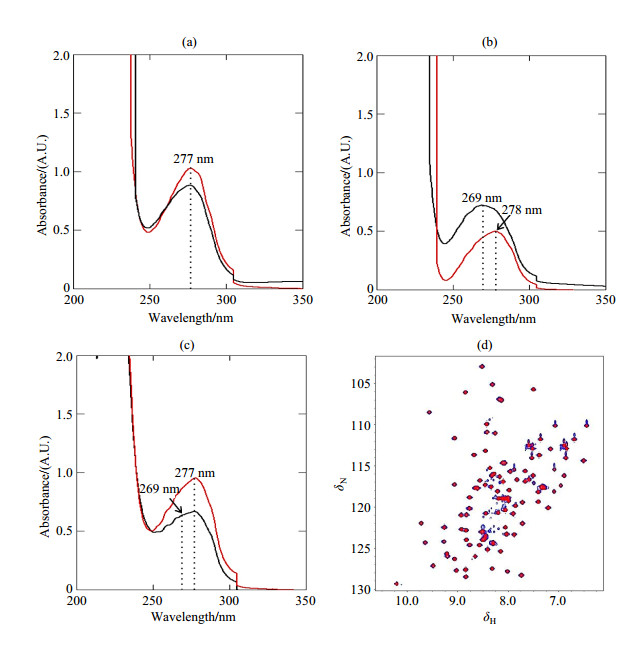
|
Fig. S1 Comparison of AtGrp7 proteins from denaturing-refolding and from regular purification by UV and NMR detection. (a) UV curves for His10 and GB1-tagged AtGrp71-90 from denaturing-refolding (red) and from regular purification (black) after the first Ni-NTA purification step. (b) and (c) are UV curves for TEV-cleaved AtGrp71-90 from denaturing-refolding (red) and from regular purification (black) after the second Ni-NTA purification step, and after size exclusion chromatography, respectively. (d) Overlapped 1H-15N HSQC spectra of free AtGrp71-90 from denaturing-refolding (blue) and from regular purification (red). All NMR data was recorded at 20 ℃. The dotted lines in Fig. (a)~(c) indicate the peak positions of the purified proteins in different stages |
To solve the problem of impurities by regular purification, a two-step denaturing-refolding protocol was tried for AtGrp71-90. In the first step, 8 mol/L urea was included in all buffers during the first Ni-NTA purification step, to denature and purify proteins from cell extracts, of which the eluted fraction had a normal UV peak center at 277 nm [Fig. S1(a)]. The eluted proteins were relatively pure in the denatured condition, and only weak bands were seen in the SDS-PAGE gel [Fig. 1(b)]. In the second step, a quick-dilution method was performed to refold AtGrp71-90. The denatured AtGrp71-90 was slowly added to a 40-fold volume of ice-cold refolding buffer with high (1 mol/L) salt concentration. No precipitate was observed, and the refolding buffer remained clear during the refolding process. For AtGrp71-90, such a refolding process by quick-dilution showed priority to the refolding process by dialysis using stepwise decreasing urea concentrations, the latter resulted in significant loss of the AtGrp7 RRM domain protein. As calculated on the unit of moles for proteins in the all purification steps, the yield of protein recovery by refolding in the quick-dilution refolding process was 81% (43.1 out of 52.9 mg) and 73% (21.6 out of 29.4 mg) (Table 1) for unlabeled and 13C, 15N-labeled AtGrp71-90, respectively. Effective TEV cleavage was observed after dialysis of TEV-included proteins overnight [Fig. 1(c)]. The resulting AtGrp71-90 from the second Ni-NTA step showed a typical UV peak for proteins, which was obviously different from that for the regular purification process [Fig. S1(b)], and suggested the effective elimination of contaminants by denaturing-refolding. Final purification by size exclusion chromatography yielded pure AtGrp71-90 with an estimated molecular weight of 12.6 kDa [Fig. 1(c)~(d)], of which the expected molecular mass is 9.8 kDa. Undoubtedly, more experiments should be performed to identify the composition of the contaminants.
| Table 1 Recombinant protein recovery yield at individual stepa |
An optional ion-exchange purification step in the denatured conditions could be applied between the first Ni-NTA and the refolding steps to further remove impurities. Eluted proteins from the first Ni-NTA step were exchanged with a low salt buffer [20 mmol/L NaPi (pH 8.0), 50 mmol/L NaCl, 0.5 mmol/L EDTA, 6 mol/L urea], and then passed through a high-performance Q column with gradient NaCl concentrations from 50 mmol/L to 2 mol/L NaCl. His10 and GB1-tagged AtGrp71-90 proteins (pI 5.68) flowed through without binding to the high-performance Q column, while most of the impurities with negative charges in the denatured condition were eluted out with increasing salt concentrations. However, the unlabeled sample with ion-exchange resulted in significant (44%) protein loss (47.0 mg loss out of 105.8 mg). Therefore, no ion-exchange was applied during the denaturing-refolding of 13C, 15N-labeled AtGrp71-90 for NMR structure determination. On the other hand, when highly pure AtGrp71-90 is required, ion exchange is still a good choice, especially if the protein amount is not the first consideration.
2.3 Verification of refolding and binding of AtGrp71-90To verify the effectiveness of AtGrp71-90 refolding, a 1H-15N HSQC spectrum was recorded for the refolded 15N-labeled protein. Peaks in 1H-15N HSQC of AtGrp71-90 were well-dispersed in a range of δ 6.0~10.4 in the 1H dimension (Fig. 2), indicating a globally folded structure of the refolded AtGrp71-90. In contrast, residue peaks of unstructured proteins were usually within δ 7.5~8.5 along the 1H dimension in the 1H-15N HSQC spectra. Success in protein refolding was further confirmed by almost fully overlapped 1H-15N HSQC spectra of the refolded AtGrp71-90 and the proteins from regular purification [Fig. S1(d)]. On the other hand, such almost same spectra lead to the question about the importance of the little amount of contaminants, implying that purification protocol of AtGrp71-90 should be further optimized.
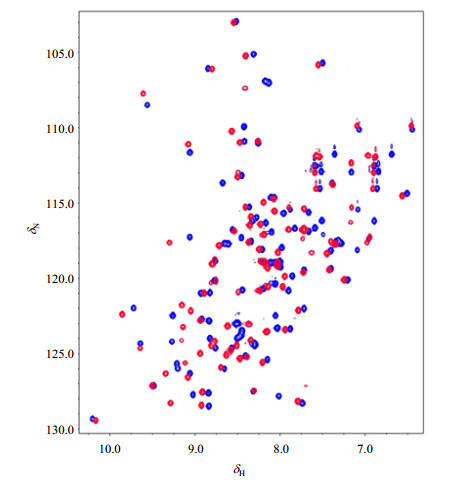
|
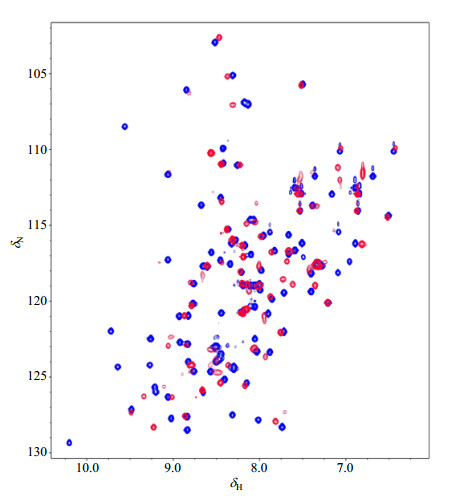
Fig. 2 Overlapped 1H-15N HSQC spectra of free AtGrp71-90 (blue) and its complex (red) with the 6-nt DNA counterpart of the RNA from 3′ UTR of AtGrp7. All NMR data was recorded at 20 ℃ |
To check whether the folded AtGrp71-90 has a proper function in substrate binding, NMR titration experiments were performed between this protein and a 6-nt DNA counterpart of the RNA from the 3′ UTR of AtGrp7[18]. Significant peak shifts in 1H-15N HSQC spectra of the complex in a molar ratio of 1:1 were observed in contrast to that of the free AtGrp71-90 (Fig. 2), indicating that the refolded AtGrp71-90 could bind to its known target. Increasing the protein to DNA molar ratio to 1:2 resulted in no change in the 1H-15N HSQC spectrum.
Quantitative analysis of the binding between the AtGrp71-90 and DNA counterparts from the 3′ UTR RNA of AtGrp7 was performed by ITC. The dissociation constant (Kd) of AtGrp71-90 and the 32-nt DNA was determined to be 0.8 μmol/L, compared to 0.1 μmol/L, as determined by fluorescence correlation spectroscopy for the full-length AtGrp7[18]. The difference in dissociation constants could be mainly ascribed to the different sizes of AtGrp7 proteins used in the two studies, and partly attributed to the different techniques. The dissociation constant decreased to 9.8 μmol/L when the critical 5′-TTCTTG -3′ sequence[18] was used (Fig. 3), suggesting that nucleotides other than the 6-nt DNA also contributed to AtGrp71-90 binding. Correspondingly, binding of AtGrp71-90 and 32-nt DNA in the molar ratio of 1:1 resulted in many disappeared or weakened peaks in the 1H-15N HSQC spectrum (Fig. S2), implying that a much stronger binding was seen in the case of AtGrp71-90 and 32-nt DNA. Our results supported the opinion that more nucleotides should be considered in the studies of DNA-protein interaction, instead of the shortest form alone, as used in structure determination.
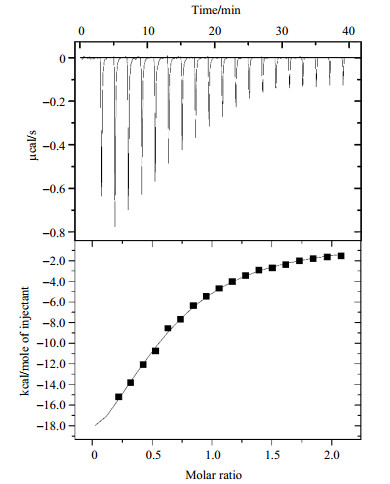
|
Fig. 3 ITC titration experiment of AtGrp71-90 and the 6-nt DNA counterpart of the RNA from 3′ UTR of AtGrp7. The concentrations of AtGrp71-90 and 6-nt DNA were 30 and 300 μmol/L, respectively. The cell volume was 200 μL and the injection volume was 40 μL, with an interval of 120 s between 20 consecutive injections. The titration experiment was performed at (25±0.1) ℃ |
|
Fig. S2 Overlapped 1H-15N HSQC spectra of free AtGrp71-90 (blue) and its complex (red) with 32-nt DNA counterpart of the RNA from 3′ UTR of AtGrp7. All NMR data was recorded at 20 ℃ |
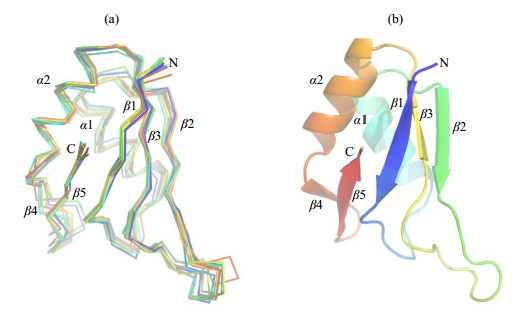
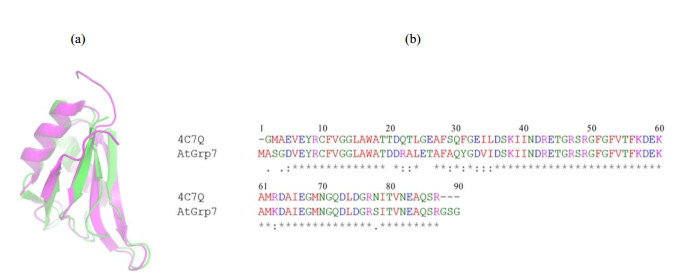
Structure models of AtGrp71-90 were constructed using chemical shifts derived from the NMR backbone assignment as input parameters by CS-Rosetta, which applied a SPARTA-based procedure to select protein fragments from the PDB database and assemble them using the Rosetta protocol. The structure models of AtGrp71-90 (Fig. 4) showed a typical RRM fold of a twisted five-stranded β-sheet together with two α-helices, in an order of βαββαββ. Analysis of ten superimposed models with the lowest energies yielded an average Cα-RMSDs of (0.07±0.01) nm against the lowest energy model. The representative structure model of AtGrp71-90 showed a high structural similarity to an RRM domain from Nicotiana tabacum with PDB code 4C7Q (Fig. S3), in accordance with their high sequence identity of 84% (and sequence similarity of 95%). Because the calculated models of AtGrp71-90 were generated by CS-Rosetta in a fragment basis, their high structural similarity to 4C7Q suggested that the models were reasonable and could be used for further binding analysis. The representative CS-Rosetta model of AtGrp71-90 was deposited at https://modelarchive.org.
|
Fig. 4 CS-Rosetta structural models of AtGrp71-90. (a) Backbone superposition of the ten structures of AtGrp71-90 with the lowest energies. (b) Structural alignment of representative CS-Rosetta model of AtGrp71-90 in ribbon representation. Structure diagrams were generated using PyMOL |
|
Fig. S3 Structure and sequence comparison between AtGrp71-90 and the Nicotiana tabacum glycine-rich RBP1 RRM domain. (a) Structural alignment of the representative CS-Rosetta model of AtGrp71-90 (green, from this study) and NMR structure (magenta, PDB code 4C7Q) of Nicotiana tabacum glycine-rich RBP1 RRM domain shown in ribbons. (b) Sequence alignment of AtGrp71-90 and Nicotiana tabacum glycine-rich RBP1 RRM domain |
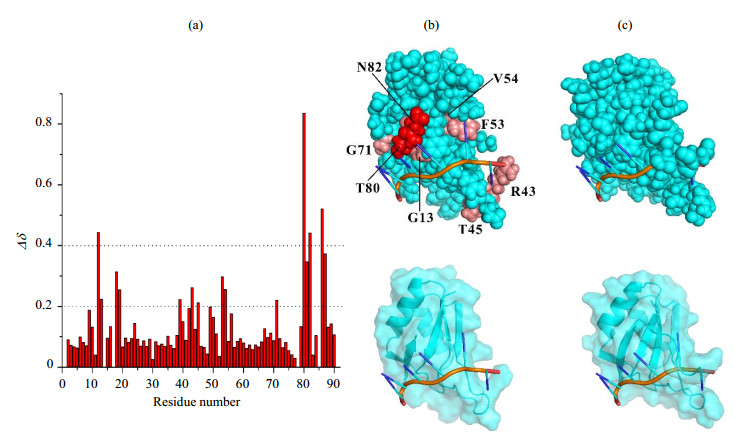
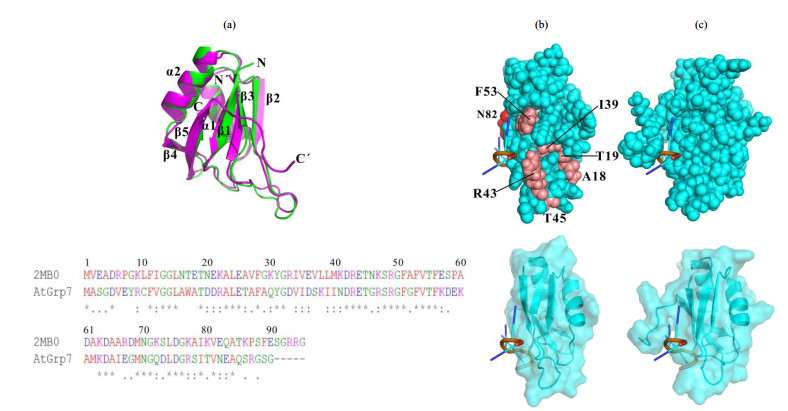
We then directly applied the 6-nt 5′-UUCUUG-3′ from 3′UTR RNA of AtGrp7 to check its binding mode on AtGrp71-90. Chemical shift perturbation mapping was obtained based on NMR backbone assignments of AtGrp71-90 with and without the 6-nt RNA. The residues on AtGrp71-90 majorly influenced by the 6-nt RNA were located at three distinct regions of the primary sequence, which were V12-A18, I39-F56, and T80-R87 in a discontinuous manner [Fig. 5(a)]. To gain a clear view of the binding mode of AtGrp71-90 with the 6-nt RNA, we generated a complex model by referring to a RRM domain (sequence identity of 44%) of hnRNP G protein in the complex with a 6-nt RNA (PDB code 2MB0). The AtGrp71-90 structure model has a relatively high structural similarity to the RRM domain of hnRNP G in complex form, with a Dali z-score[28] of 11.9 [Fig. S4(a)]. Meanwhile, its structural similarity to the aforementioned RRM domain from the Nicotiana tabacum glycine-rich RBP1 (PDB code 4C7Q) is 13.2. In general, the complex model of AtGrp71-90 and its 6-nt RNA substrate matched well with the chemical shift perturbation data [top and bottom of Fig. 5(b)]. A couple of residues (A18, T19, T45) experienced relatively large chemical shift changes while they are far away from the RNA binding surface on the model structure, which were attributed to local environment changes of the corresponding residues in loops. The main residues of AtGrp71-90 involved in RNA binding (with a chemical shift change of δ ≥ 0.4) were V12, T80, N82, and S86. Surprisingly, the residues around T80-R87 had large chemical shift changes due to RNA binding, which were unstructured in the free AtGrp71-90, as indicated by the structure model calculation. Structure analysis of the similar RRM domain of hnRNP G in complex form suggested that the residues around T80-R87 of AtGrp71-90 may experience large conformational changes after RNA binding [Fig. S4(b) and (c)], which were missed in the structure models. On the other hand, R49 in AtGrp71-90, which was a key residue for RNA binding[29], did not show significant chemical shift changes after RNA binding [Fig. 5(a)]. Since our ITC data showed that the dissociation constant of the AtGrp71-90 R49Q mutant and the corresponding 6-nt DNA decreased by one order of magnitude compared to that of the wild-type AtGrp71-90 (data not shown), it is more likely that the backbone amides of R49 are less sensitive to RNA/DNA binding, and a high-resolution structure is required to elucidate the molecular details of AtGrp71-90/RNA binding.
|
Fig. 5 Binding analysis of AtGrp71-90 with 6-nt RNA from 3′ UTR of AtGrp7. (a) Chemical shift differences of residues between free AtGrp71-90 and its complex with 6-nt RNA, which were calculated using Eq. (1). (b) Complex model of AtGrp71-90 and 6-nt RNA in sphere mode (top) and in surface-cartoon combined mode (bottom). Residues on AtGrp71-90 with chemical shift differences in the range of δ 0.2~0.4 are shown in salmon, and those in the range of δ ≥ 0.4 are shown in red. The residues around T80-R87 of AtGrp71-90 with large chemical shift changes after RNA binding were missed in the structure models. (c) Complex of the RRM domain of hnRNP G protein and a 6-nt RNA (PDB code 2MB0) in sphere mode (top) and in surface-cartoon combined mode (bottom). In Fig. (b) and (c), the proteins are shown in cyan, and the RNAs are shown in red, which indicates the backbone. Structure diagrams were generated using PyMOL |
|
Fig. S4 Structure and binding comparison between AtGrp71-90 and the RRM domain of hnRNP G protein. (a) Structural alignment of the representative CS-Rosetta model of AtGrp71-90 (green, from this study) and NMR structure (magenta, PDB code 2MB0) of the RRM domain of hnRNP G protein shown in ribbons (top) and their sequence alignment (bottom). (b) Complex model of AtGrp71-90 and 6-nt RNA in sphere mode (top) and in surface-cartoon combined mode (bottom). Residues on AtGrp71-90 with chemical shift differences in the range of δ 0.2~0.4 are shown in salmon, and those in the range of δ ≥ 0.4 are shown in red. The residues around T80-R87 of AtGrp71-90 with large chemical shift changes due to RNA binding were missed in the structure models. (c) Complex of the RRM domain of hnRNP G protein and a 6-nt RNA (PDB code 2MB0) in sphere mode (top) and in surface-cartoon combined mode (bottom). In Fig. (b) and (c), structures are shown by rotating them by 90 degrees with respect to those in Fig. (b) and (c), and the proteins are shown in cyan, and the RNAs are shown in red, indicating the backbone |
AtGrp7RRM, encoding the RRM domain AtGrp71-90 of Arabidopsis thaliana, was cloned into a modified pET16b vector in which GB1-fusion could improve the yield, solubility, and stability of AtGrp71-90. By using a quick-dilution refolding procedure, a satisfactory yield of protein was obtained for NMR structure determination; the residual contaminants in the purified protein were also avoided. But still, current results suggest that more experiments have to be performed to identify the composition of the contaminants and to optimize the purification protocol of AtGrp71-90. The structure of the AtGrp7 RRM domain was fully recovered by refolding, as supported by its finger-print 1H-15N HSQC spectrum and CS-Rosetta model structures. ITC and NMR titration experiments further confirmed that the RRM domain of AtGrp71-90 had the right function of RNA/DNA binding. In future studies, we aim to obtain atomic level three-dimensional NMR structures of the AtGrp7 RRM domain and its complex with target RNA to understand its function as an RNA/DNA chaperone.
| [1] | HEINTZEN C, NATER M, APEL K, et al. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana[J]. Proc Natl Acad Sci U S A, 1997, 94(16): 8515-8520. DOI: 10.1073/pnas.94.16.8515. |
| [2] | STREITNER C, HENNIN L, KORNELI C, et al. Global transcript profiling of transgenic plants constitutively overexpressing the RNA-binding protein AtGRP7[J]. BMC Plant Biol, 2010, 10: 221. DOI: 10.1186/1471-2229-10-221. |
| [3] | MEYER K, KÖSTER T, NOLTE C, et al. Adaptation of iCLIP to plants determines the binding landscape of the clock regulated RNA-binding protein AtGRP7[J]. Genome Biol, 2017, 18(1): 204. |
| [4] | STREITNER C, KÖSTER T, SIMPSON C G, et al. An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana[J]. Nucleic Acids Res, 2012, 40(22): 11240-11255. DOI: 10.1093/nar/gks873. |
| [5] | KÖSTER T, MEYER K, WEINHOLDT C, et al. Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis[J]. Nucleic Acids Res, 2014, 42(15): 9925-9936. DOI: 10.1093/nar/gku716. |
| [6] | STREITNER C, DANISMAN S, WEHRLE F, et al. The small glycine-rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana[J]. Plant J, 2008, 56(2): 239-250. DOI: 10.1111/j.1365-313X.2008.03591.x. |
| [7] | KIM J S, JUNG H J, LEE H J, et al. Glycine-rich RNA-binding protein7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana[J]. Plant J, 2008, 55(3): 455-466. DOI: 10.1111/tpj.2008.55.issue-3. |
| [8] | FU Z Q, GUO M, JEONG B R, et al. A type Ⅲ effector ADP-ribosylates RNA-binding proteins and quells plant immunity[J]. Nature, 2007, 447(7142): 284-288. DOI: 10.1038/nature05737. |
| [9] | JEONG B R, LIN Y, JOE A, et al. Structure function analysis of an ADP-ribosyltransferase type Ⅲ effector and its RNA-binding target in plant immunity[J]. J Biol Chem, 2011, 286(50): 43272-43281. DOI: 10.1074/jbc.M111.290122. |
| [10] | NICAISE V, JOE A, JEONG B R, et al. Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7[J]. EMBO J, 2013, 32(5): 701-712. DOI: 10.1038/emboj.2013.15. |
| [11] | HACKMANN C, KORNELI C, KUTYNIOK M, et al. Salicylic acid-dependent and -independent impact of an RNA-binding protein on plant immunity[J]. Plant Cell Environ, 2014, 37(3): 696-706. DOI: 10.1111/pce.2014.37.issue-3. |
| [12] | KIM J S, PARK S J, KWAK K J, et al. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli[J]. Nucleic Acids Res, 2007, 35(2): 506-516. |
| [13] | WANG S C, LIANG D, SHI S G, et al. Isolation and characterization of a novel drought responsive gene encoding a glycine-rich RNA-binding protein in Malus prunifolia (Willd.) Borkh[J]. Plant Mol Biol Rep, 2011, 29(1): 125-134. DOI: 10.1007/s11105-010-0221-1. |
| [14] | CAO S Q, JIANG L, SONG S Y, et al. AtGrp7 is involved in the regulation of abscisic acid and stress responses in Arabidopsis[J]. Cell Mol Biol Lett, 2006, 11(4): 526-535. |
| [15] | LEDER V, LUMMER M, TEGELER K, et al. Mutational definition of binding requirements of an hnRNP-like protein in Arabidopsis using fluorescence correlation spectroscopy[J]. Biochem Biophys Res Commun, 2014, 453(1): 69-74. DOI: 10.1016/j.bbrc.2014.09.056. |
| [16] | TRIPET B P, MASON K E, EILER B J, et al. Structural and biochemical analysis of the Hordeum vulgare L. HvGR-RBP1 protein, a glycine-rich rna-binding protein involved in the regulation of barley plant development and stress response[J]. Biochemistry, 2014, 53(50): 7945-7960. DOI: 10.1021/bi5007223. |
| [17] | FRANCO-ECHEVARRIA, GONZÁLEZ-POLO N, ZORRILLA S, et al. The structure of transcription termination factor Nrd1 reveals an original mode for GUAA recognition[J]. Nucleic Acids Res, 2017, 45(17): 10293-10305. DOI: 10.1093/nar/gkx685. |
| [18] | SCHüTTPELZ M, SCHöNING J C, DOOSE S, et al. Changes in conformational dynamics of mRNA upon AtGRP7 binding studied by Fluorescence correlation spectroscopy[J]. J Am Chem Soc, 2008, 130: 9507-9513. DOI: 10.1021/ja801994z. |
| [19] | DELAGLIO F, GRZESIEK S, VUISTER G W, et al. NMRPipe:a multidimensional spectral processing system based on UNIX pipes[J]. J Biomol NMR, 1995, 6(3): 277-293. |
| [20] | JOHNSON B A, BLEVINS R A. NMRView:a computer program for the visualization and analysis of NMR data[J]. J Biomol NMR, 1994, 4(5): 603-614. DOI: 10.1007/BF00404272. |
| [21] | LANGE O F, ROSSI P, SGOURAKIS N G, et al. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples[J]. Proc Natl Acad Sci U S A, 2012, 109(27): 10873-10878. DOI: 10.1073/pnas.1203013109. |
| [22] | SHEN Y, VEMON R, BAKER D, et al. De novo protein structure generation from incomplete chemical shift assignments[J]. J Biomol NMR, 2009, 43(2): 63-78. DOI: 10.1007/s10858-008-9288-5. |
| [23] | SHEN Y, LANGE O, DELAGLIO F, et al. Consistent blind protein structure generation from NMR chemical shift data[J]. Proc Natl Acad Sci U S A, 2008, 105(12): 4685-4690. DOI: 10.1073/pnas.0800256105. |
| [24] | FINN R D, COGGILL P, EBERHARDT R Y, et al. The Pfam protein families database:towards a more sustainable future[J]. Nucleic Acids Res, 2016, 44(D1): D279-D285. DOI: 10.1093/nar/gkv1344. |
| [25] | KHAN F, DANIËLS M A, FOLKERS G E, et al. Structural basis of nucleic acid binding by Nicotiana tabacum glycine-rich RNA-binding protein:implications for its RNA chaperone function[J]. Nucleic Acids Res, 2014, 42(13): 8705-8718. DOI: 10.1093/nar/gku468. |
| [26] | HUTH J R, BEWLEY C A, JACKSON B M, et al. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR[J]. Protein Sci, 1997, 6(11): 2359-2364. |
| [27] | QIAO X Y, LIU Y, LUO L T, et al. Effects of naturally occurring charged mutations on the structure, stability, and binding of the Pin1 WW domain[J]. Biochem Biophys Res Commun, 2017, 487(2): 470-476. DOI: 10.1016/j.bbrc.2017.04.093. |
| [28] | HOLM L, ROSENSTRÖM P. Dali server:conservation mapping in 3D[J]. Nucleic Acids Res, 2010, 38: W545-W549. DOI: 10.1093/nar/gkq366. |
| [29] | SCHÖNING J C, STREITNER C, PAGE D R, et al. Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation[J]. Plant J, 2007, 52(6): 1119-1130. DOI: 10.1111/tpj.2007.52.issue-6. |
 2019, Vol. 36
2019, Vol. 36 
